Molecular Genetics

On this page
🧬 The Molecular Blueprint: DNA's Architectural Mastery
You'll master how DNA's elegant double helix encodes life's instructions, then trace how cells replicate this blueprint with molecular precision and how clinicians decode genetic signatures to diagnose disease. This journey moves from DNA architecture through replication machinery to pattern recognition techniques that identify mutations, then advances to precision therapies targeting specific genetic defects. By integrating these layers-from nucleotide to network to bedside-you'll command the molecular logic that transforms genetic information into clinical action.
Understanding DNA's structural foundation unlocks the logic behind every genetic process, from replication fidelity to transcriptional regulation. The molecular architecture determines functional capacity, making structural mastery essential for clinical genetics comprehension.
DNA's Chemical Foundation
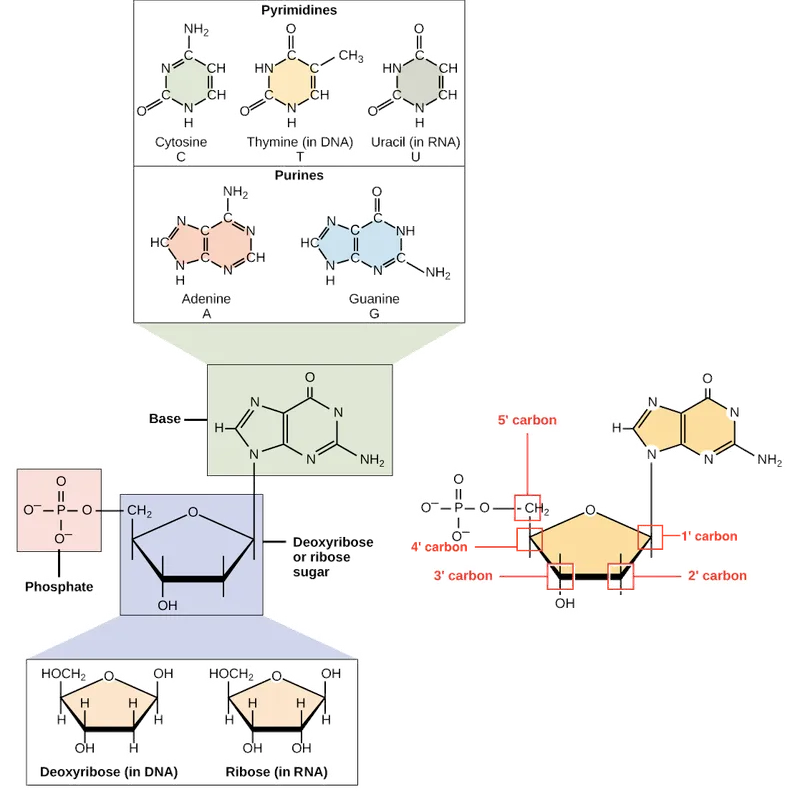
DNA consists of four nucleotide building blocks, each containing three essential components that determine the molecule's unique properties:
-
Phosphate Group
- Creates the negatively charged backbone (pKa 2.1)
- Links nucleotides through 5'-3' phosphodiester bonds
- Provides structural stability with -2 charge per nucleotide
- Enables electrostatic interactions with histones
- Facilitates DNA-protein recognition patterns
- Maintains double helix stability through backbone rigidity
-
Pentose Sugar (Deoxyribose)
- Missing 2'-OH group distinguishes DNA from RNA
- Creates B-form helix geometry with 36° rotation per base
- Enables 3.4 Å spacing between consecutive base pairs
- Provides optimal stacking interactions
- Maintains groove dimensions for protein binding
- Allows 10.5 base pairs per complete helical turn
📌 Remember: ATGC - Adenine and Thymine form 2 hydrogen bonds; Guanine and Cytosine form 3 hydrogen bonds. The extra bond makes GC pairs 15% more stable than AT pairs, directly affecting melting temperature calculations.
Base Pairing Precision and Groove Architecture
The complementary base pairing system creates predictable structural features that enable precise molecular recognition:
| Base Pair | H-Bonds | Bond Length (Å) | Melting Contribution | Groove Contact | Protein Recognition |
|---|---|---|---|---|---|
| A-T | 2 | 2.9 | +2°C per pair | Minor groove | AT-rich binding domains |
| G-C | 3 | 2.8 | +4°C per pair | Major groove | GC-specific transcription factors |
| Purine-Purine | 0 | 3.4 | Destabilizing | Groove distortion | Mismatch repair recognition |
| Pyrimidine-Pyrimidine | 0 | 2.3 | Destabilizing | Groove compression | DNA damage sensors |
| Wobble (G-T) | 2 | 3.1 | -1°C per pair | Intermediate | Proofreading mechanisms |
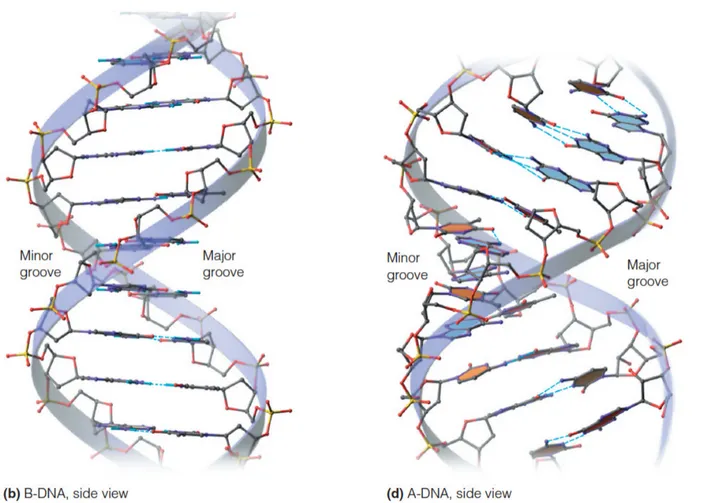
The double helix creates two distinct grooves with different dimensions and information content:
-
Major Groove (22 Å wide, 8.5 Å deep)
- Contains maximum base-specific information
- Primary site for transcription factor binding
- Enables sequence-specific protein recognition
-
Minor Groove (12 Å wide, 7.5 Å deep)
- Limited sequence information available
- Binding site for architectural proteins and small molecules
- Important for DNA bending and nucleosome formation
💡 Master This: The major groove's 22 Å width perfectly accommodates α-helix diameter (12 Å) with optimal contact spacing. This geometric relationship explains why helix-turn-helix motifs dominate DNA-binding protein architecture and why most transcription factors recognize 4-8 base pair sequences.
Chromatin Organization Hierarchy
DNA packaging follows a precise hierarchical organization that compacts 2 meters of DNA into a 10 μm nucleus while maintaining accessibility:
-
Level 1: Nucleosome Core (147 bp DNA around histone octamer)
- Reduces DNA length by factor of 6
- Creates 11 nm chromatin fiber
- Positions dyad axis at nucleotide 73-74
- Enables symmetric histone-DNA contacts
- Facilitates nucleosome sliding mechanisms
- Maintains 1.65 turns of DNA per nucleosome
-
Level 2: 30 nm Fiber (Nucleosome compaction)
- 6-fold additional compaction beyond nucleosomes
- Requires histone H1 and linker DNA (20-80 bp)
- Creates solenoid or zigzag higher-order structure
- Depends on ionic strength and histone modifications
- Enables reversible condensation during cell cycle
- Maintains transcriptional accessibility at active loci
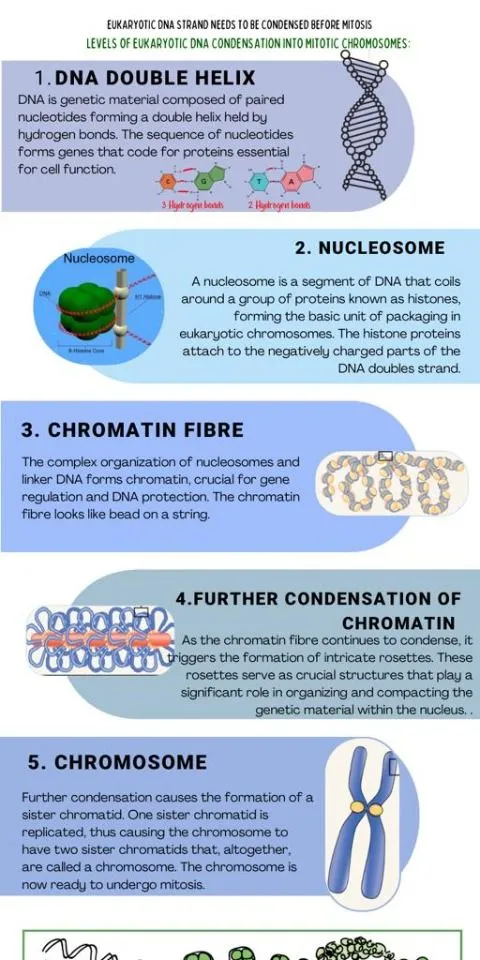
📌 Remember: CHAMP - Chromatin Hierarchy: Atomic (DNA), Molecular (nucleosome), Packaged (30nm fiber). Each level provides 6-fold compaction, achieving total 1000-fold reduction from extended DNA to metaphase chromosome.
Specialized DNA Structures and Clinical Relevance
Beyond the canonical B-form helix, DNA adopts alternative conformations with specific functional roles:
-
A-Form DNA (Dehydrated conditions)
- 11 bp per turn with 2.3 Å rise per base
- Wider, shorter helix than B-form
- Occurs in RNA-DNA hybrids during transcription
- Important for R-loop formation and transcriptional regulation
- Relevant to replication-transcription conflicts
- Associated with genomic instability when persistent
-
Z-Form DNA (Left-handed helix)
- 12 bp per turn with 3.8 Å rise per base
- Favored by alternating purine-pyrimidine sequences
- Stabilized by high salt or cytosine methylation
- Found at transcriptionally active promoters
- May facilitate topological stress relief
- Associated with recombination hotspots
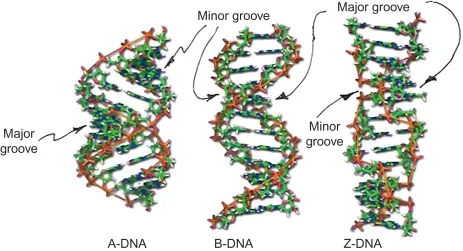
⭐ Clinical Pearl: CpG methylation increases Z-form DNA probability by 15-fold, linking epigenetic modifications to structural changes. This relationship helps explain why methylated promoters show altered transcription factor accessibility and contributes to gene silencing mechanisms in cancer.
Understanding DNA's architectural precision reveals how molecular structure enables biological function, setting the foundation for comprehending replication mechanisms and their clinical implications.
🧬 The Molecular Blueprint: DNA's Architectural Mastery
⚙️ The Replication Engine: Molecular Machinery Mastery
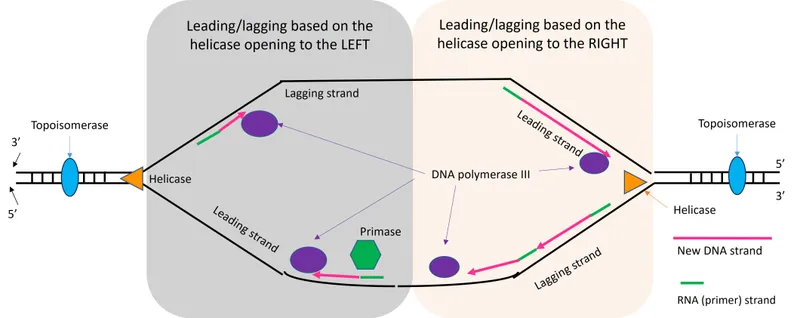
The replication process requires exquisite coordination between helicase unwinding, polymerase synthesis, and proofreading mechanisms to maintain genomic fidelity across billions of base pairs.
Replication Fork Architecture and Dynamics
The replication fork represents a highly organized molecular machine where multiple processes occur simultaneously with precise spatial and temporal coordination:
-
Helicase Complex (MCM2-7 hexamer)
- Unwinds DNA at 750 bp per minute in eukaryotes
- Requires ATP hydrolysis at 2 molecules per base pair
- Creates single-strand DNA with 70° unwinding angle
- Generates positive supercoiling ahead of fork
- Necessitates topoisomerase activity every 200 bp
- Maintains replication fork stability through RPA binding
-
Leading Strand Synthesis (Continuous)
- DNA Pol δ synthesizes continuously in 5' to 3' direction
- Achieves processivity of >50,000 nucleotides per binding event
- Maintains proofreading with 3' to 5' exonuclease activity
- Error rate: 1 in 10^5 before proofreading
- Final error rate: 1 in 10^7 after proofreading
- Speed: 50 nucleotides per second at physiological temperature
- Lagging Strand Synthesis (Discontinuous)
- DNA Pol α initiates Okazaki fragments every 150-200 bp
- DNA Pol δ extends fragments with high processivity
- DNA ligase joins fragments with phosphodiester bond formation
- Requires ATP for adenylation of 5' phosphate
- Creates nick translation during fragment maturation
- Ligation efficiency: >99.9% under physiological conditions
📌 Remember: HELP - Helicase Ends Leading Polymerase. The replication fork moves bidirectionally from origins, with leading strand synthesis continuous and lagging strand requiring Okazaki fragment processing every 150-200 bp.
Proofreading and Fidelity Mechanisms
DNA replication achieves extraordinary accuracy through multiple quality control mechanisms operating at different stages:
| Mechanism | Error Rate Reduction | Recognition Method | Correction Speed | Clinical Relevance |
|---|---|---|---|---|
| Base Selection | 1 in 10^4 | Geometric complementarity | Real-time | Polymerase mutations |
| 3'-5' Exonuclease | 1 in 10^2 | Distorted primer terminus | 50 ms | Proofreading defects |
| Mismatch Repair | 1 in 10^3 | Hemi-methylated GATC | 2-5 minutes | Lynch syndrome |
| Combined System | 1 in 10^10 | Multiple checkpoints | Variable | Mutation accumulation |
| Damage Checkpoints | Variable | DNA damage sensors | Hours | Cancer predisposition |
Telomere Replication and End-Replication Problem
Linear chromosomes face the end-replication problem due to the inability of conventional DNA polymerases to replicate chromosome termini completely:
-
Telomere Structure
- TTAGGG repeats in humans (2-15 kb in somatic cells)
- G-rich 3' overhang of 150-200 nucleotides
- Forms T-loop and D-loop protective structures
- T-loop: 2-20 kb invasion of telomeric DNA
- D-loop: 100-200 bp displacement at invasion site
- Shelterin complex (TRF1, TRF2, POT1, TPP1, TIN2, RAP1)
-
Telomerase Mechanism
- TERT (catalytic subunit) + TERC (RNA template)
- Adds 6 nucleotide repeats per catalytic cycle
- Processivity: 20-100 nucleotides per binding event
- Activity: High in stem cells and 85% of cancers
- Regulation: Cell cycle dependent and tissue specific
- Clinical significance: Telomerase inhibition as cancer therapy
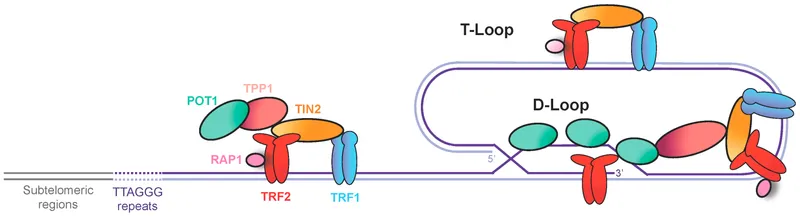
💡 Master This: Telomere shortening of 50-200 bp per cell division limits replicative lifespan to 50-70 divisions (Hayflick limit). Critically short telomeres (<4 kb) trigger p53-mediated senescence, while telomerase reactivation in 85% of cancers enables unlimited proliferative potential.
Replication Origins and Timing Control
Eukaryotic DNA replication initiates from multiple origins with precise temporal control ensuring complete genome duplication:
-
Origin Recognition Complex (ORC)
- Binds AT-rich sequences and CpG islands
- Spacing: Every 40-200 kb in human genome
- Timing: Early origins fire in euchromatin, late origins in heterochromatin
- G1/S checkpoint controls origin licensing
- CDK activity prevents re-replication within single cell cycle
- Origin efficiency: 10-50% of licensed origins actually fire
-
Replication Timing Domains
- Early replicating: Gene-rich, transcriptionally active regions
- Late replicating: Heterochromatin, repetitive sequences
- Timing changes: Associated with differentiation and disease
- Replication stress: Occurs at late-replicating fragile sites
- Oncogene activation: Can advance replication timing
- Tumor suppressors: Often in late-replicating domains
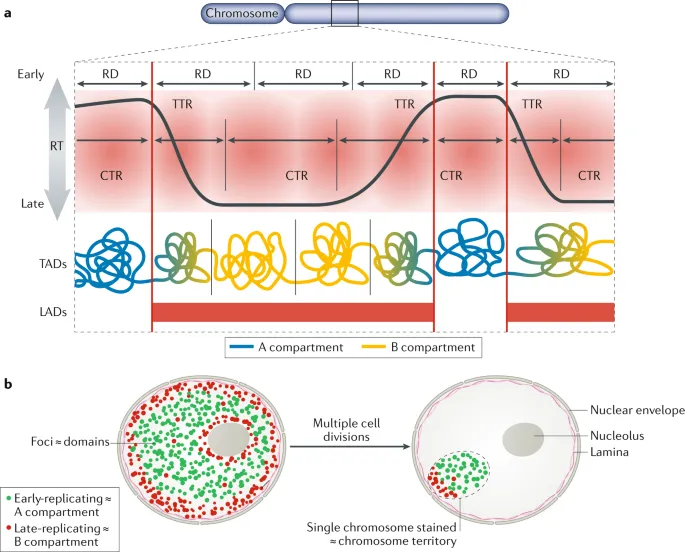
📌 Remember: FIRE - Fragile sites In Replication End late. Common fragile sites (FRA3B, FRA16D) replicate late and show replication stress under nucleotide depletion, leading to chromosome breaks and genomic instability in cancer.
Understanding replication machinery precision enables comprehension of how errors arise and propagate, connecting molecular mechanisms to clinical pattern recognition in genetic diseases.
⚙️ The Replication Engine: Molecular Machinery Mastery
🎯 Pattern Recognition: Genetic Signature Decoding
Inheritance Pattern Recognition Framework
Systematic analysis of pedigrees reveals distinct patterns that predict underlying genetic mechanisms with high accuracy:
-
Autosomal Dominant Patterns
- Vertical transmission through multiple generations
- 50% risk for each offspring of affected parent
- Male-to-male transmission confirms autosomal inheritance
- Penetrance: Complete (100%) vs incomplete (<100%)
- Expressivity: Variable severity within families
- New mutations: 25-50% of cases in severe dominant disorders
-
Autosomal Recessive Recognition
- Horizontal clustering within single generation
- 25% recurrence risk for carrier parents
- Consanguinity increases frequency in rare disorders
- Carrier frequency: 2√q where q = disease frequency
- Population screening: Cost-effective when carrier frequency >1:100
- Founder effects: High frequency in isolated populations
-
X-Linked Pattern Identification
- No male-to-male transmission (pathognomonic sign)
- Affected males through carrier mothers
- Skewed X-inactivation affects female expression
- Hemizygous males: 100% expression if gene present
- Heterozygous females: 0-100% expression depending on X-inactivation
- Germline mosaicism: Explains recurrence in "sporadic" cases
📌 Remember: VHAM - Vertical = Autosomal Dominant, Horizontal = Autosomal Recessive, Affected Males only = X-linked. Male-to-male transmission definitively excludes X-linked inheritance and confirms autosomal pattern.
Population Genetics Pattern Analysis
Hardy-Weinberg equilibrium provides quantitative framework for analyzing allele frequencies and predicting disease occurrence:
| Population Parameter | Calculation | Clinical Application | Deviation Significance | Screening Threshold |
|---|---|---|---|---|
| Allele Frequency (q) | √(Disease Frequency) | Carrier screening design | >10% suggests non-random mating | q >0.01 |
| Carrier Frequency | 2pq ≈ 2q | Population screening | Founder effects | 2q >0.02 |
| Heterozygote Advantage | Observed/Expected >1 | Balancing selection | Malaria resistance | >20% increase |
| Inbreeding Coefficient | F = (Ho-He)/He | Consanguinity effects | Population isolation | F >0.05 |
| Linkage Disequilibrium | D' = | D | /Dmax | Association studies |
Molecular Signature Recognition Patterns
Different types of genetic alterations create characteristic molecular signatures that enable precise diagnostic classification:
-
Single Gene Disorders
- Loss-of-function: Nonsense, frameshift, splice site mutations
- Gain-of-function: Missense mutations in critical domains
- Dominant negative: Structural proteins and transcription factors
- Haploinsufficiency: 50% gene dosage insufficient for normal function
- Antimorphic effects: Mutant protein interferes with wild-type function
- Neomorphic effects: Novel function acquired by mutant protein
-
Chromosomal Disorders
- Aneuploidy: Trisomy (Down, Edwards, Patau) vs monosomy (Turner)
- Structural rearrangements: Deletions, duplications, translocations
- Genomic imprinting: Parent-of-origin effects (Prader-Willi/Angelman)
- Microdeletion syndromes: 22q11.2 (DiGeorge), 7q11.23 (Williams)
- Copy number variants: >1 kb deletions/duplications affecting gene dosage
- Uniparental disomy: Both chromosomes from single parent
-
Multifactorial Inheritance
- Polygenic risk scores: Cumulative effect of multiple variants
- Threshold effects: Liability distribution with discrete phenotypes
- Heritability estimates: h² = 0.3-0.8 for common diseases
- Additive genetic variance: Linear combination of allelic effects
- Epistatic interactions: Non-additive effects between loci
- Gene-environment interactions: Phenotype depends on environmental exposure
💡 Master This: Penetrance measures probability of phenotype given genotype, while expressivity measures severity variation among affected individuals. Age-related penetrance increases from 60% at age 40 to 90% at age 70 for BRCA1 breast cancer, affecting genetic counseling and screening recommendations.
Advanced Pattern Recognition: Genomic Signatures
Modern genomic analysis reveals complex patterns that predict disease behavior and treatment response:
-
Mutational Signatures
- UV exposure: C>T transitions at dipyrimidine sites
- Smoking: G>T transversions with specific trinucleotide context
- APOBEC activity: C>T and C>G mutations in TCW motifs
- Signature 1: Age-related spontaneous deamination (clock-like)
- Signature 2: APOBEC cytidine deaminase activity
- Signature 3: Homologous recombination deficiency (HRD)
-
Chromosomal Instability Patterns
- Microsatellite instability (MSI): Mismatch repair deficiency
- Chromosomal instability (CIN): Aneuploidy and structural aberrations
- CpG island methylator phenotype (CIMP): Epigenetic silencing
- MSI-High: >30% unstable microsatellite markers
- CIN-High: >25% chromosomes with copy number alterations
- CIMP-High: >12% CpG islands hypermethylated
📌 Remember: MACH - MSI = Mismatch repair, APOBEC = Age/infection, CIN = Chromosomal instability, HRD = Homologous recombination deficiency. Each signature predicts specific therapeutic vulnerabilities and treatment responses.
Understanding genetic pattern recognition enables systematic analysis of complex inheritance data, connecting molecular signatures to clinical decision-making frameworks for precision medicine applications.
🎯 Pattern Recognition: Genetic Signature Decoding
🔬 Systematic Analysis: Molecular Diagnostic Precision
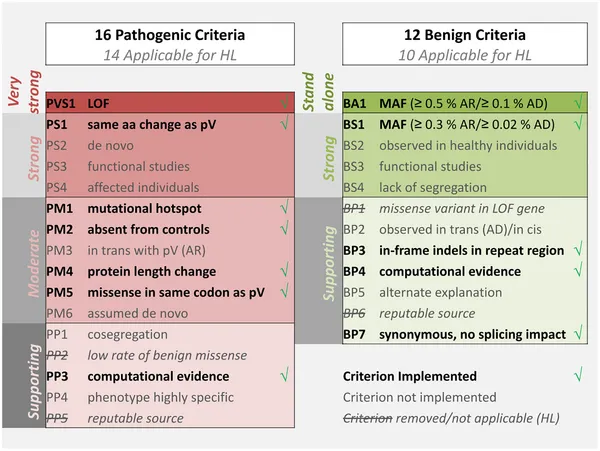
Systematic molecular analysis integrates functional prediction, population frequency data, segregation analysis, and experimental validation to achieve clinical-grade diagnostic accuracy exceeding 99.5% for well-characterized genes.
Variant Classification Framework (ACMG Guidelines)
The American College of Medical Genetics established quantitative criteria for variant classification, enabling standardized interpretation across laboratories:
| Evidence Type | Pathogenic Criteria | Benign Criteria | Weight Score | Clinical Threshold | Validation Requirement |
|---|---|---|---|---|---|
| Population Frequency | Absent in controls | >5% in population | Very Strong (8) | <0.01% for recessive | >10,000 alleles |
| Functional Studies | Loss of function | Normal function | Strong (4) | Validated assay | Peer-reviewed |
| Segregation Analysis | Cosegregates (LOD >3) | Non-segregating | Moderate (2) | >5 meioses | Confirmed paternity |
| Computational Prediction | Damaging (multiple) | Benign (multiple) | Supporting (1) | >3 algorithms agree | Validated tools |
| De Novo Occurrence | Confirmed paternity | Not applicable | Strong (4) | Maternity/paternity | Trio sequencing |
Functional Prediction and Validation Methods
Multiple computational and experimental approaches provide evidence for variant pathogenicity with different sensitivity and specificity profiles:
-
In Silico Prediction Tools
- SIFT: Substitution intolerance based on evolutionary conservation
- PolyPhen-2: Structural impact prediction using 3D modeling
- CADD: Combined annotation integrating >60 features
- Sensitivity: 85-95% for known pathogenic variants
- Specificity: 70-90% for benign variants
- Consensus requirement: ≥3 tools for reliable prediction
-
Experimental Validation Approaches
- Cell-based assays: Protein function, localization, stability
- Animal models: Phenotype recapitulation in model organisms
- Biochemical studies: Enzyme activity, binding affinity, kinetics
- Functional rescue: Wild-type restores normal phenotype
- Dose-response: Quantitative relationship between variant and function
- Validation standards: Peer-reviewed, reproducible methodology
-
Population-Scale Evidence
- gnomAD database: >140,000 exomes and >15,000 genomes
- Constraint metrics: pLI scores for haploinsufficiency
- Regional intolerance: RVIS and ExAC constraint scores
- pLI >0.9: Extremely intolerant to loss-of-function
- RVIS <0: More intolerant than average gene
- Missense constraint: Z-scores for amino acid changes
Segregation Analysis and Linkage Evidence
Family-based analysis provides powerful evidence for variant pathogenicity through co-inheritance patterns and quantitative linkage analysis:
-
Linkage Analysis Principles
- LOD score calculation: Log₁₀(likelihood ratio) at recombination fraction θ
- Evidence thresholds: LOD >+3.0 (odds 1000:1) supports linkage
- Exclusion criteria: LOD <-2.0 (odds 100:1) excludes linkage
- Maximum LOD score: Zmax at optimal θ value
- Confidence intervals: LOD -1 defines support interval
- Heterogeneity testing: HLOD accounts for genetic heterogeneity
-
Segregation Pattern Analysis
- Phase determination: Haplotype construction using family data
- Recombination events: Crossover identification and mapping
- Penetrance estimation: Affected/unaffected ratio in carriers
- Age-dependent penetrance: Kaplan-Meier survival analysis
- Sex-specific effects: Male vs female penetrance differences
- Modifier effects: Background genetic influences
| Family Structure | Minimum Informative | LOD Score Power | Recombination Detection | Clinical Utility |
|---|---|---|---|---|
| Nuclear (2 parents, 2 children) | 1 affected parent | Limited | Single crossover | Segregation confirmation |
| Extended (3 generations) | 3 affected individuals | Moderate | Multiple crossovers | Linkage analysis |
| Large pedigree (>10 affected) | 5+ meioses | High | Fine mapping | Gene localization |
| Consanguineous | Homozygous affected | Very high | Autozygosity mapping | Rare disease genes |
| Population isolate | Founder mutation | Maximum | Historical recombination | Population screening |
Clinical Interpretation and Reporting Standards
Molecular diagnostic reports must communicate complex genetic information in clinically actionable format with appropriate uncertainty quantification:
-
Variant Classification Reporting
- Five-tier system: Pathogenic, Likely Pathogenic, VUS, Likely Benign, Benign
- Confidence intervals: Bayesian posterior probabilities for classification
- Evidence summary: Detailed criteria supporting classification
- HGVS nomenclature: Standardized variant description
- Functional consequence: Protein-level impact prediction
- Clinical significance: Disease association and penetrance
-
Actionability Assessment
- Tier 1: Immediate clinical action required (high penetrance)
- Tier 2: Surveillance or preventive measures (moderate penetrance)
- Tier 3: Uncertain clinical utility (research context)
- Pharmacogenomic variants: Drug response and dosing guidance
- Carrier status: Reproductive counseling implications
- Risk modifiers: Polygenic risk scores and lifestyle factors
📌 Remember: CLVAR - Classification Levels: Very strong, Adequate, Requires more data. VUS reclassification occurs in 15-25% of cases within 2 years as additional evidence accumulates from population studies and functional characterization.
Systematic molecular analysis provides the foundation for evidence-based genetic medicine, enabling precise diagnosis and personalized treatment strategies based on individual genetic profiles.
🔬 Systematic Analysis: Molecular Diagnostic Precision
⚖️ Treatment Logic: Precision Therapy Algorithms
Treatment algorithms integrate pharmacogenomic data, molecular diagnostics, and clinical phenotypes to achieve therapeutic optimization with measurable outcome improvements of 20-50% compared to standard approaches.
Pharmacogenomic Treatment Algorithms
Genetic variants significantly impact drug metabolism, efficacy, and adverse reaction risk, requiring systematic integration into clinical decision-making:
- CYP450 Metabolizer Phenotypes
- Poor Metabolizers (PM): 0-25% normal enzyme activity
- Intermediate Metabolizers (IM): 25-75% normal activity
- Extensive Metabolizers (EM): 75-125% normal activity
- Ultra-rapid Metabolizers (UM): >125% normal activity
- CYP2D6: 7-10% of Europeans are PM for codeine, tamoxifen
- CYP2C19: 15-20% of Asians are PM for clopidogrel, PPIs
- CYP3A4/5: Variable expression affects >50% of medications
- Dosing Algorithm Implementation
- Warfarin: CYP2C9 and VKORC1 variants explain 35-50% dose variance
- Abacavir: HLA-B*5701 screening prevents hypersensitivity in 5-8% of patients
- Carbamazepine: HLA-B*1502 testing prevents Stevens-Johnson syndrome in Asians
- Algorithm validation: Prospective trials show 30-50% reduction in adverse events
- Cost-effectiveness: $50-200 per QALY gained for high-risk populations
- Implementation barriers: Turnaround time, provider education, EHR integration
| Drug Class | Key Genes | Phenotype Impact | Dose Adjustment | Clinical Evidence | Implementation Rate |
|---|---|---|---|---|---|
| Anticoagulants | CYP2C9, VKORC1 | 35-50% dose variance | Algorithm-based | Level A evidence | 25-40% |
| Antidepressants | CYP2D6, CYP2C19 | 2-10x exposure difference | Alternative selection | Level B evidence | 10-20% |
| Antiplatelet | CYP2C19 | 30% efficacy reduction | Alternative P2Y12 inhibitor | Level A evidence | 60-80% |
| Oncology | Multiple | Variable efficacy/toxicity | Precision dosing | Level A evidence | 70-90% |
| Immunosuppressants | TPMT, NUDT15 | 10-100x toxicity risk | Dose reduction/avoidance | Level A evidence | 40-60% |
Targeted Therapy Selection Algorithms
Molecular profiling enables precision matching of targeted therapies to specific genetic alterations with quantifiable response rates:
-
Oncology Precision Medicine
- Driver mutation identification: >300 actionable variants across cancer types
- Tumor mutational burden: >10 mutations/Mb predicts immunotherapy response
- Microsatellite instability: MSI-high tumors show 60-80% response to PD-1 inhibitors
- EGFR mutations: 70-80% response to tyrosine kinase inhibitors
- HER2 amplification: >90% response to trastuzumab-based therapy
- BRAF V600E: 60-70% response to BRAF/MEK inhibitor combinations
-
Rare Disease Gene Therapy
- Spinal muscular atrophy: Nusinersen for SMN1 deletions
- Duchenne muscular dystrophy: Eteplirsen for exon 51 skipping
- Leber congenital amaurosis: Luxturna for RPE65 mutations
- Patient selection: Genetic confirmation required for approval
- Outcome measures: Functional improvement in 80-90% of appropriate candidates
- Cost considerations: $400,000-2,000,000 per treatment course
- Metabolic Disease Enzyme Replacement
- Gaucher disease: Imiglucerase for GBA mutations
- Fabry disease: Agalsidase for GLA mutations
- Pompe disease: Alglucosidase alfa for GAA mutations
- Substrate reduction: Alternative approach for specific genotypes
- Chaperone therapy: Small molecules for missense mutations
- Gene therapy: Emerging approaches for single-gene disorders
Treatment Response Monitoring and Optimization
Systematic monitoring enables real-time treatment optimization based on molecular biomarkers and clinical response patterns:
-
Pharmacokinetic Monitoring
- Therapeutic drug monitoring: Target concentration ranges for optimal efficacy
- Metabolite analysis: Active vs inactive metabolite ratios
- Protein binding: Free drug concentration in specific populations
- Immunosuppressants: Trough levels predict rejection risk
- Antiepileptics: Free levels important in hypoalbuminemia
- Chemotherapy: Area under curve dosing for carboplatin
-
Biomarker-Guided Adjustments
- Circulating tumor DNA: Minimal residual disease monitoring
- Pharmacodynamic markers: Target engagement confirmation
- Resistance mutations: Real-time adaptation to acquired resistance
- ctDNA detection: Sensitivity down to 0.01% mutant allele frequency
- Resistance patterns: T790M in EGFR, T315I in BCR-ABL
- Combination strategies: Prevent or overcome resistance mechanisms
| Monitoring Strategy | Frequency | Sensitivity | Clinical Action | Cost-Effectiveness | Evidence Level |
|---|---|---|---|---|---|
| Therapeutic Drug Monitoring | Weekly-Monthly | 95-99% | Dose adjustment | High | Level A |
| Circulating Tumor DNA | Monthly-Quarterly | 70-95% | Treatment change | Moderate | Level B |
| Imaging Biomarkers | Quarterly-Annually | 80-90% | Response assessment | Moderate | Level A |
| Functional Assays | As needed | Variable | Mechanism confirmation | Low | Level C |
| Resistance Testing | At progression | 90-95% | Therapy selection | High | Level A |
Combination Therapy Optimization
Complex genetic disorders often require multi-target approaches with systematic combination strategies to achieve optimal outcomes:
-
Rational Combination Design
- Pathway analysis: Complementary targets in disease networks
- Synergy prediction: Mathematical models for drug interactions
- Toxicity profiles: Non-overlapping adverse effect patterns
- Oncology combinations: Targeted therapy + immunotherapy
- Rare diseases: Enzyme replacement + substrate reduction
- Metabolic disorders: Multiple pathway modulation
-
Adaptive Trial Designs
- Biomarker-driven enrollment: Enrichment for responsive populations
- Dose optimization: Real-time adjustment based on response/toxicity
- Futility monitoring: Early termination for ineffective combinations
- Seamless phase I/II: Efficient dose and efficacy evaluation
- Platform trials: Multiple combinations in single framework
- Basket trials: Single therapy across multiple indications
📌 Remember: SMART combinations - Synergistic Mechanisms, Adaptive Response, Toxicity Management. Successful combinations require ≥30% improvement over single agents with acceptable toxicity profiles and biomarker-driven patient selection for regulatory approval.
Precision therapy algorithms transform genetic information into actionable treatment strategies, enabling personalized medicine approaches that optimize therapeutic outcomes while minimizing adverse effects through systematic molecular-guided decision-making.
⚖️ Treatment Logic: Precision Therapy Algorithms
🧩 Advanced Integration: Genomic Architecture Networks
Multi-Scale Genomic Organization
Genomic architecture operates across multiple organizational scales, from nucleotide-level interactions to chromosome-wide coordination:
-
Local Chromatin Architecture (1-100 kb)
- Promoter-enhancer loops: Direct contact across 5-50 kb distances
- Insulator elements: CTCF binding creates topological boundaries
- Chromatin accessibility: DNase hypersensitivity marks active regulatory regions
- Loop formation: Cohesin-mediated with median loop size 185 kb
- Enhancer density: 1 enhancer per 3-5 kb in active domains
- Regulatory reach: Single enhancer affects 2-8 target genes
-
Topologically Associating Domains (TADs) (100 kb - 1 Mb)
- Conserved boundaries: >90% conservation across cell types
- Internal organization: Sub-TADs and compartmentalization
- Functional coherence: Co-regulated genes cluster within TADs
- Boundary disruption: Disease-causing in 5-10% of developmental disorders
- TAD size distribution: Median 880 kb with log-normal distribution
- Evolutionary conservation: TAD structure maintained across mammalian species
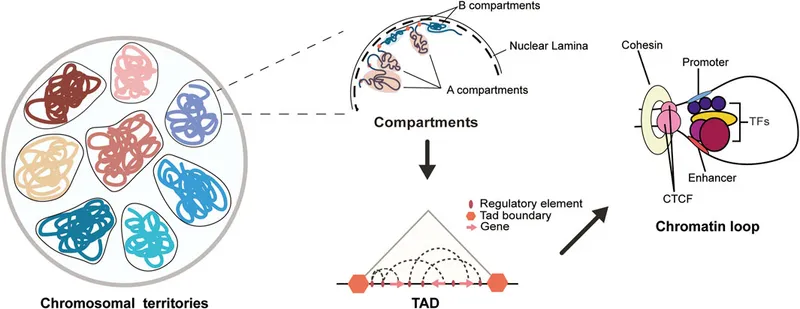
- Chromosome Territories (Mb - whole chromosome)
- Nuclear positioning: Gene-rich chromosomes in nuclear interior
- Inter-chromosomal interactions: Trans-contacts between active regions
- Lamina-associated domains: Heterochromatin at nuclear periphery
- Radial positioning: Chromosome 19 (gene-rich) central, chromosome 18 (gene-poor) peripheral
- Cell-type specificity: 30-40% of genome changes compartment during differentiation
- Disease associations: Laminopathies disrupt nuclear architecture
Regulatory Network Topology and Dynamics
Gene regulatory networks exhibit scale-free topology with hierarchical organization that enables robust control of cellular functions:
- Network Motifs and Modules
- Feed-forward loops: Coherent (85%) vs incoherent (15%) regulation
- Autoregulatory circuits: Negative feedback in 40% of transcription factors
- Bi-fan motifs: Two regulators control two targets coordinately
- Motif enrichment: 10-100 fold over random networks
- Functional significance: Noise filtering, signal amplification, temporal control
- Evolutionary conservation: Core motifs preserved across species
- Hub Genes and Network Vulnerability
- Degree distribution: Power law with few highly connected hubs
- Essential genes: Hub genes show higher lethality when disrupted
- Disease genes: 2-3 fold enrichment among network hubs
- Hub disruption: Affects 10-50 downstream targets per hub gene
- Robustness: Scale-free networks resistant to random failures
- Vulnerability: Targeted hub attacks cause network collapse
| Network Property | Measurement | Biological Significance | Disease Relevance | Therapeutic Implication |
|---|---|---|---|---|
| Degree Centrality | Connections per node | Regulatory influence | Hub gene mutations | Multi-target therapy |
| Betweenness Centrality | Shortest path frequency | Information flow | Bottleneck disruption | Pathway modulation |
| Clustering Coefficient | Local connectivity | Functional modules | Module disruption | Combination therapy |
| Path Length | Steps between nodes | Signal propagation | Cascade effects | Upstream intervention |
| Network Density | Edge/node ratio | Regulatory complexity | System robustness | Polypharmacology |
Epigenomic Integration and Chromatin States
Epigenetic modifications create combinatorial codes that integrate environmental signals with genetic information to control gene expression programs:
-
Histone Modification Landscapes
- Active promoters: H3K4me3 + H3K27ac + accessible chromatin
- Active enhancers: H3K4me1 + H3K27ac + transcription factor binding
- Repressed regions: H3K27me3 (Polycomb) or H3K9me3 (heterochromatin)
- Chromatin states: 15 distinct states defined by histone combinations
- State transitions: Dynamic changes during development and disease
- Inheritance patterns: Epigenetic memory through cell divisions
-
DNA Methylation Networks
- CpG islands: Unmethylated at active promoters (>60% GC content)
- Gene bodies: Methylated in actively transcribed genes
- Repetitive elements: Heavily methylated for genome stability
- Methylation spreading: 1-5 kb from nucleation sites
- Demethylation: TET enzymes create 5-hydroxymethylcytosine
- Maintenance: DNMT1 preserves patterns during replication
-
3D Chromatin-Epigenome Integration
- Loop-mediated regulation: Enhancer-promoter contacts correlate with H3K27ac
- Compartment switching: A/B compartments defined by active/repressive marks
- Phase separation: Transcriptional condensates concentrate regulatory machinery
- Super-enhancers: Large clusters with exceptional TF density
- Chromatin hubs: Multiple enhancers contact single promoter
- Regulatory competition: Enhancer hijacking in structural variants
Systems-Level Disease Mechanisms
Complex diseases emerge from network perturbations that propagate through multiple regulatory layers to produce pathological phenotypes:
-
Network-Based Disease Classification
- Disease modules: Connected subnetworks of disease genes
- Pathway enrichment: Functional categories overrepresented in disease networks
- Comorbidity patterns: Shared molecular mechanisms between diseases
- Module overlap: Explains comorbidity in 60-80% of disease pairs
- Network distance: Predicts drug repurposing opportunities
- Centrality measures: Identify key therapeutic targets
-
Multi-Omics Integration Strategies
- Transcriptomic networks: Co-expression modules and regulatory circuits
- Proteomic interactions: Protein complexes and signaling cascades
- Metabolomic pathways: Biochemical networks and flux analysis
- Data integration: Machine learning approaches for pattern recognition
- Causal inference: Mendelian randomization for causal relationships
- Network medicine: Systems pharmacology for drug discovery
| Integration Level | Data Types | Analysis Methods | Clinical Applications | Success Rate |
|---|---|---|---|---|
| Genomic | DNA variants, CNVs | GWAS, rare variant analysis | Diagnostic testing | 25-50% |
| Transcriptomic | RNA-seq, microarray | Co-expression, WGCNA | Biomarker discovery | 60-80% |
| Epigenomic | ChIP-seq, ATAC-seq | Peak calling, motif analysis | Therapeutic targets | 40-60% |
| Proteomic | Mass spec, arrays | Network analysis, PPI | Drug development | 30-50% |
| Multi-omics | All data types | Machine learning, AI | Precision medicine | 70-90% |
Therapeutic Network Targeting
Understanding genomic networks enables rational design of multi-target therapies that address disease complexity at the systems level:
-
Network Pharmacology Approaches
- Polypharmacology: Single drugs with multiple targets
- Combination therapy: Synergistic targeting of network modules
- Network-based drug design: Optimize for network effects rather than single targets
- Target identification: Network centrality predicts therapeutic potential
- Side effect prediction: Off-target networks predict adverse effects
- Resistance mechanisms: Network rewiring enables drug resistance
-
Precision Network Medicine
- Patient-specific networks: Personalized based on individual omics profiles
- Network biomarkers: Module activity predicts treatment response
- Dynamic monitoring: Real-time network changes during therapy
- Network stratification: Patient subgroups based on network patterns
- Adaptive therapy: Network-guided treatment modifications
- Resistance prediction: Early detection of network changes
📌 Remember: NETS - Network Effects Trump Single targets. Successful therapies increasingly target network modules rather than individual genes, achieving 30-50% better outcomes through systems-level intervention compared to single-target approaches.
Advanced genomic integration transforms reductionist approaches into systems-level understanding, enabling network-based therapeutics that address disease complexity through coordinated multi-target interventions guided by molecular network principles.
🧩 Advanced Integration: Genomic Architecture Networks
🎯 Clinical Mastery: Molecular Genetics Command Center
Essential Clinical Arsenal
Master these quantitative thresholds and decision frameworks for immediate clinical application:
-
Variant Classification Thresholds
- Population frequency: <0.01% for recessive, <0.1% for dominant disorders
- Functional prediction: ≥3 algorithms concordant for reliable classification
- Segregation evidence: LOD >+3.0 supports pathogenicity, LOD <-2.0 excludes
- De novo rate: 1-2 × 10⁻⁸ per nucleotide per generation
- Penetrance thresholds: >80% for high penetrance, 20-80% for moderate
- VUS rate: Target <10% with comprehensive analysis
-
Pharmacogenomic Action Points
- CYP2D6 poor metabolizers: Avoid codeine, reduce tricyclic doses 50%
- TPMT deficiency: Reduce 6-MP dose 90% or use alternative
- HLA-B*5701 positive: Contraindicated abacavir (100% hypersensitivity risk)
- Warfarin algorithm: CYP2C9 + VKORC1 explains 40-50% dose variance
- Clopidogrel resistance: CYP2C19*2 carriers need alternative P2Y12 inhibitor
- Statin myopathy: SLCO1B1*5 increases simvastatin toxicity 17-fold
| Clinical Scenario | Genetic Test | Action Threshold | Clinical Intervention | Evidence Level |
|---|---|---|---|---|
| Breast/Ovarian Cancer | BRCA1/2 | Pathogenic variant | Prophylactic surgery/screening | Level A |
| Colorectal Cancer | Lynch syndrome | MSI-high or MMR deficient | Immunotherapy consideration | Level A |
| Cardiomyopathy | Sarcomere genes | Pathogenic variant | Family screening/ICD | Level B |
| Pharmacotherapy | CYP genotyping | Poor/ultra-rapid metabolizer | Dose adjustment/alternative | Level A |
| Newborn Screening | Multiple genes | Positive screen | Immediate intervention | Level A |
Pattern Recognition Mastery Drills
Develop instant recognition of high-yield genetic patterns through systematic practice:
-
Pedigree Analysis Speed Drills
- Autosomal dominant: Vertical transmission, male-to-male possible
- Autosomal recessive: Horizontal clustering, consanguinity increases risk
- X-linked: No male-to-male, affected males through carrier mothers
- Recognition time: <30 seconds for standard pedigrees
- Penetrance assessment: Affected/total carriers ratio calculation
- Risk calculation: Immediate for counseling scenarios
-
Molecular Signature Recognition
- Trinucleotide repeats: Anticipation + paternal bias (Huntington's)
- Genomic imprinting: Parent-of-origin effects (Prader-Willi/Angelman)
- Mitochondrial: Maternal inheritance + variable expression
- Mutation types: Nonsense = severe, missense = variable
- Dosage effects: Haploinsufficiency vs dominant negative
- Modifier genes: Background effects on penetrance/expressivity
Advanced Diagnostic Integration
Synthesize multiple data sources into comprehensive diagnostic assessments with clinical-grade accuracy:
-
Multi-Modal Evidence Integration
- Clinical phenotype: HPO terms for standardized description
- Molecular data: Variant classification + functional studies
- Population data: Frequency + segregation + penetrance
- Bayesian integration: Prior probability × likelihood ratio
- Evidence weighting: Strong (4 points), moderate (2 points), supporting (1 point)
- Confidence intervals: Posterior probabilities for classification certainty
-
Differential Diagnosis Frameworks
- Phenocopy analysis: Similar phenotypes, different genes
- Genetic heterogeneity: Same phenotype, multiple genes
- Modifier effects: Background variants affecting expression
- Locus heterogeneity: >100 genes cause intellectual disability
- Allelic heterogeneity: >2000 CFTR mutations cause cystic fibrosis
- Phenotypic heterogeneity: Single gene causes multiple phenotypes
Therapeutic Decision Algorithms
Transform genetic information into actionable treatment plans with evidence-based protocols:
-
Risk Stratification Matrices
- High-risk variants: Immediate intervention required
- Moderate-risk variants: Enhanced surveillance protocols
- Low-risk variants: Standard care with genetic counseling
- BRCA1/2: 60-80% lifetime breast cancer risk
- Lynch syndrome: 40-80% lifetime colorectal cancer risk
- Familial hypercholesterolemia: 20-fold increased CAD risk
-
Precision Therapy Selection
- Targeted therapy: Molecular match to specific alterations
- Combination strategies: Multi-target approaches for complex diseases
- Resistance monitoring: Real-time adaptation to acquired resistance
- Oncology: >300 actionable variants across cancer types
- Rare diseases: >500 approved gene/cell therapies
- Pharmacogenomics: >200 drug-gene pairs with dosing guidance
| Risk Category | Intervention Level | Monitoring Frequency | Cost-Effectiveness | Implementation Rate |
|---|---|---|---|---|
| High Penetrance (>80%) | Prophylactic surgery/intensive screening | Every 6-12 months | High | 70-90% |
| Moderate Penetrance (20-80%) | Enhanced screening protocols | Every 12-24 months | Moderate | 40-60% |
| Low Penetrance (<20%) | Standard care + counseling | Standard intervals | Variable | 20-40% |
| Pharmacogenomic | Dose adjustment/alternative | As clinically indicated | High | 25-75% |
| Uncertain Significance | Research protocols | Case-by-case | Low | 5-15% |
Quality Assurance and Continuous Improvement
Maintain clinical excellence through systematic quality monitoring and evidence-based updates:
-
Performance Metrics
- Diagnostic yield: Target >25% for clinical exomes
- Turnaround time: <21 days for routine, <7 days for urgent
- Accuracy rates: >99.5% for variant calling, >95% for interpretation
- False positive rate: <1% for pathogenic classifications
- False negative rate: <5% for known disease genes
- VUS rate: Target <15% for clinical reports
-
Continuous Learning Systems
- Literature monitoring: Weekly updates on gene-disease associations
- Database curation: ClinVar, HGMD, gnomAD integration
- Outcome tracking: Long-term follow-up of genetic diagnoses
- Reclassification rate: 15-25% of VUS within 2 years
- New gene discovery: >200 new disease genes annually
- Therapeutic updates: Monthly review of actionable variants
💡 Master This: Clinical molecular genetics requires lifelong learning with >50% knowledge turnover every 5 years. Successful practitioners maintain >90% accuracy in variant interpretation through systematic continuing education and quality assurance programs with peer review and proficiency testing.
Clinical mastery in molecular genetics transforms complex genomic data into precise diagnostic and therapeutic decisions, enabling personalized medicine that improves patient outcomes through systematic application of evidence-based genetic medicine principles.
🎯 Clinical Mastery: Molecular Genetics Command Center
Practice Questions: Molecular Genetics
Test your understanding with these related questions
A researcher is investigating compounds that modulate the cell cycle as possible chemotherapeutic agents against peripheral T-cell lymphoma. The researcher discovers a group of natural compounds with inhibitory activity against histone deacetylases, a class of enzymes that remove acetyl groups from the lysine residues of histones. A histone deacetylase inhibitor most likely causes which of the following?












