Plasmids and mobile genetic elements US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Plasmids and mobile genetic elements. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Plasmids and mobile genetic elements US Medical PG Question 1: The surgical equipment used during a craniectomy is sterilized using pressurized steam at 121°C for 15 minutes. Reuse of these instruments can cause transmission of which of the following pathogens?
- A. Non-enveloped viruses
- B. Sporulating bacteria
- C. Prions (Correct Answer)
- D. Enveloped viruses
- E. Yeasts
Plasmids and mobile genetic elements Explanation: ***Prions***
- Prions are **abnormally folded proteins** that are highly resistant to standard sterilization methods like steam autoclaving at 121°C, making them a risk for transmission through reused surgical instruments.
- They cause transmissible spongiform encephalopathies (TSEs) like **Creutzfeldt-Jakob disease**, where even trace amounts can be highly infectious.
*Non-enveloped viruses*
- Non-enveloped viruses are generally **more resistant to heat and disinfectants** than enveloped viruses but are typically inactivated by recommended steam sterilization protocols.
- Standard autoclaving conditions are effective in destroying most non-enveloped viruses.
*Sporulating bacteria*
- **Bacterial spores**, such as those from *Clostridium* or *Bacillus*, are known for their high resistance to heat and chemicals, but are usually **inactivated by steam sterilization at 121°C** for 15 minutes.
- This method is specifically designed to kill bacterial spores effectively.
*Enveloped viruses*
- Enveloped viruses are the **least resistant to heat and chemical disinfectants** due to their lipid envelope.
- They are readily **inactivated by standard steam sterilization** at 121°C.
*Yeasts*
- **Yeasts** are eukaryotic microorganisms that are typically **susceptible to heat sterilization**.
- They are effectively killed by typical steam autoclaving conditions used for surgical instruments.
Plasmids and mobile genetic elements US Medical PG Question 2: A codon is an mRNA sequence consisting of 3 nucleotides that codes for an amino acid. Each position can be made up of any 4 nucleotides (A, U, G, C); therefore, there are a total of 64 (4 x 4 x 4) different codons that can be created but they only code for 20 amino acids. This is explained by the wobble phenomenon. One codon for leucine is CUU, which of the following can be another codon coding for leucine?
- A. CUA (Correct Answer)
- B. CCC
- C. UAA
- D. CCA
- E. AUG
Plasmids and mobile genetic elements Explanation: ***CUA***
- The **wobble hypothesis** allows for non-standard base pairing at the **third position** of the codon.
- Since CUU codes for leucine, a change in the third base to **A (CUA)** can often still code for the same amino acid due to degeneracy of the genetic code.
*CCC*
- This codon codes for **proline**, not leucine.
- A change in the **second letter** of the codon almost always results in a different amino acid.
*UAA*
- This is one of the **stop codons** (UAA, UAG, UGA), which signals the termination of translation.
- It does not code for any amino acid.
*CCA*
- This codon codes for **proline**, not leucine.
- Changing the first or second nucleotide typically results in a different amino acid.
*AUG*
- This codon codes for **methionine** and also serves as the **start codon** for protein synthesis.
- It does not code for leucine.
Plasmids and mobile genetic elements US Medical PG Question 3: An outbreak of diphtheria has occurred for the third time in a decade in a small village in South Africa. Diphtheria is endemic to the area with many healthy villagers colonized with different bacterial strains. Vaccine distribution in this area is difficult due to treacherous terrain. A team of doctors is sent to the region to conduct a health campaign. Toxigenic strains of C. diphtheria are isolated from symptomatic patients. Which of the following best explains the initial emergence of a pathogenic strain causing such outbreaks?
- A. Infection with a lytic phage
- B. Conjugation between the toxigenic and non-toxigenic strains of C. diphtheriae
- C. Suppression of lysogenic cycle
- D. Lysogenic conversion (Correct Answer)
- E. Presence of naked DNA in the environment
Plasmids and mobile genetic elements Explanation: ***Lysogenic conversion***
- **Lysogenic conversion** occurs when a temperate bacteriophage infects a bacterium and integrates its DNA into the bacterial genome, carrying genes that confer new properties, such as **toxin production**.
- The **diphtheria toxin** gene is encoded by the *tox* gene carried by the **beta-phage**, which integrates into *Corynebacterium diphtheriae* via lysogeny, converting a non-pathogenic strain into a pathogenic one.
*Infection with a lytic phage*
- A **lytic phage** infects a bacterium, replicates rapidly, and then lyses the host cell, releasing new phage particles; it typically does not integrate into the host genome to confer new stable properties like toxin production.
- Lytic phages are primarily responsible for bacterial destruction, not for conferring new stable virulence factors like the **diphtheria toxin**.
*Conjugation between the toxigenic and non-toxigenic strains of C. diphtheriae*
- **Conjugation** involves the direct transfer of genetic material via a pilus between bacteria, usually involving plasmids. While it can transfer virulence factors, the **diphtheria toxin gene** is chromosomally integrated via a **phage**, not typically transferred through conjugation in this manner.
- *C. diphtheriae* toxin production is specifically associated with the presence of the **toxin gene from a lysogenic bacteriophage**, not plasmid-mediated transfer between strains.
*Suppression of lysogenic cycle*
- **Suppression of the lysogenic cycle** means the phage exits the dormant lysogenic state and enters the lytic cycle, leading to host cell lysis. This would not explain the *initial emergence* or stable acquisition of toxin production.
- If the lysogenic cycle were suppressed, the integrated phage (and thus the **toxin gene**) might be lost or the host cell destroyed, rather than stably expressing a new pathogenic trait.
*Presence of naked DNA in the environment*
- The presence of **naked DNA** in the environment leads to **transformation**, where bacteria take up free DNA from their surroundings. While this can transfer genes, the **diphtheria toxin gene** is specifically introduced into *C. diphtheriae* by a **lysogenic bacteriophage**, not typically by free environmental DNA.
- Transformation is a mechanism for acquiring genetic material, but the origin and mechanism of acquisition for the **diphtheria toxin gene** are well-established as phage-mediated.
Plasmids and mobile genetic elements US Medical PG Question 4: DNA replication is a highly complex process where replication occurs on both strands of DNA. On the leading strand of DNA, replication occurs uninterrupted, but on the lagging strand, replication is interrupted and occurs in fragments called Okazaki fragments. These fragments need to be joined, which of the following enzymes is involved in the penultimate step before ligation can occur?
- A. DNA gyrase
- B. DNA ligase
- C. DNA helicase
- D. DNA polymerase I (Correct Answer)
- E. DNA polymerase III
Plasmids and mobile genetic elements Explanation: **DNA polymerase I**
- **DNA polymerase I** plays a crucial role in removing the **RNA primers** from the Okazaki fragments on the lagging strand.
- After primer removal, it fills the resulting gaps with **deoxyribonucleotides** before DNA ligase seals the nicks.
*DNA gyrase*
- **DNA gyrase** (a type of **topoisomerase**) is involved in relieving **supercoiling** ahead of the replication fork.
- It does not directly participate in the joining of Okazaki fragments, but rather in maintaining DNA topology during replication.
*DNA ligase*
- **DNA ligase** is responsible for the **final sealing** of the nicks between adjacent Okazaki fragments.
- It forms a **phosphodiester bond** between the 3'-hydroxyl end of one fragment and the 5'-phosphate end of the next, following primer removal and gap filling.
*DNA helicase*
- **DNA helicase** unwinds the double-stranded DNA helix, separating the two strands at the **replication fork**.
- This enzyme is essential for initiating replication but does not participate in processing Okazaki fragments.
*DNA polymerase III*
- **DNA polymerase III** is the primary enzyme responsible for the **elongation of new DNA strands** in both leading and lagging strand synthesis.
- It synthesizes the actual Okazaki fragments but does not directly remove primers or fill the gaps.
Plasmids and mobile genetic elements US Medical PG Question 5: A 64-year-old female with type 2 diabetes mellitus comes to the physician because of a 1-week history of painful red swelling on her left thigh. Examination shows a 3- x 4-cm, tender, fluctuant mass. Incision and drainage of the abscess are performed. Culture of the abscess fluid grows gram-positive, coagulase-positive cocci that are resistant to oxacillin. Which of the following best describes the mechanism of resistance of the causal organism to oxacillin?
- A. Degradation of the antibiotic
- B. Decreased uptake of the antibiotic
- C. Decreased activation of the antibiotic
- D. Altered target of the antibiotic (Correct Answer)
- E. Acetylation of the antibiotic
Plasmids and mobile genetic elements Explanation: ***Altered target of the antibiotic***
- The organism described (gram-positive, coagulase-positive cocci, oxacillin-resistant) is **methicillin-resistant *Staphylococcus aureus* (MRSA)**.
- MRSA achieves oxacillin (and other beta-lactam) resistance by acquiring the ***mecA* gene**, which encodes for a **modified penicillin-binding protein (PBP2a)** with reduced affinity for beta-lactam antibiotics.
*Degradation of the antibiotic*
- This mechanism, primarily through the production of **beta-lactamase enzymes**, can degrade beta-lactam antibiotics.
- While *Staphylococcus aureus* can produce beta-lactamases, oxacillin (a **penicillinase-resistant penicillin**) is specifically engineered to be stable against these enzymes.
*Decreased uptake of the antibiotic*
- Reduced permeability of the bacterial cell wall can lead to decreased uptake, a mechanism more commonly associated with **gram-negative bacteria** due to their outer membrane.
- This is not the primary mechanism of resistance for MRSA to oxacillin.
*Decreased activation of the antibiotic*
- Some antibiotics are prodrugs that require activation by bacterial enzymes, and resistance can arise from mutations affecting this activation.
- Oxacillin is active in its administered form and does not require bacterial activation.
*Acetylation of the antibiotic*
- **Enzymatic modification**, such as acetylation, adenylylation, or phosphorylation, is a common mechanism of resistance, particularly against **aminoglycoside antibiotics**.
- This specific mechanism is not responsible for oxacillin resistance in MRSA.
Plasmids and mobile genetic elements US Medical PG Question 6: A scientist is studying the mechanisms by which bacteria become resistant to antibiotics. She begins by obtaining a culture of vancomycin-resistant Enterococcus faecalis and conducts replicate plating experiments. In these experiments, colonies are inoculated onto a membrane and smeared on 2 separate plates, 1 containing vancomycin and the other with no antibiotics. She finds that all of the bacterial colonies are vancomycin resistant because they grow on both plates. She then maintains the bacteria in liquid culture without vancomycin while she performs her other studies. Fifteen generations of bacteria later, she conducts replicate plating experiments again and finds that 20% of the colonies are now sensitive to vancomycin. Which of the following mechanisms is the most likely explanation for why these colonies have become vancomycin sensitive?
- A. Point mutation
- B. Gain of function mutation
- C. Viral infection
- D. Plasmid loss (Correct Answer)
- E. Loss of function mutation
Plasmids and mobile genetic elements Explanation: ***Plasmid loss***
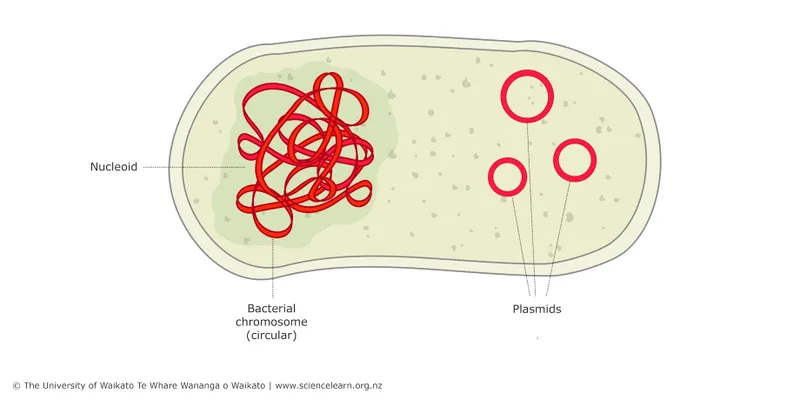
- The initial **vancomycin resistance** in *Enterococcus faecalis* is often mediated by genes located on **plasmids**, which are extrachromosomal DNA.
- In the absence of selective pressure (vancomycin), bacteria that lose the plasmid (and thus the resistance genes) have a **growth advantage** over those that retain the energetically costly plasmid, leading to an increase in sensitive colonies over generations.
*Point mutation*
- A **point mutation** typically involves a change in a single nucleotide and could lead to loss of resistance if it occurred in a gene conferring resistance.
- However, since there was no selective pressure for loss of resistance, it is less likely that 20% of the population would acquire such a specific point mutation to revert resistance.
*Gain of function mutation*
- A **gain of function mutation** would imply that the bacteria acquired a *new* advantageous trait, not the *loss* of resistance.
- This type of mutation would not explain why some colonies became sensitive to vancomycin after the drug was removed.
*Viral infection*
- **Viral infection** (bacteriophages) can transfer genes through transduction or cause bacterial lysis, but it's not the primary mechanism for a widespread reversion of resistance in the absence of antibiotic pressure.
- It would not explain the observed increase in vancomycin-sensitive colonies due to evolutionary pressure.
*Loss of function mutation*
- While a **loss of function mutation** in a gene conferring resistance could lead to sensitivity, it's generally less likely to explain a 20% shift without selective pressure than **plasmid loss**.
- Plasmids are often unstable and are easily lost in the absence of selection, whereas a specific gene mutation causing loss of function would need to arise and become prevalent in the population.
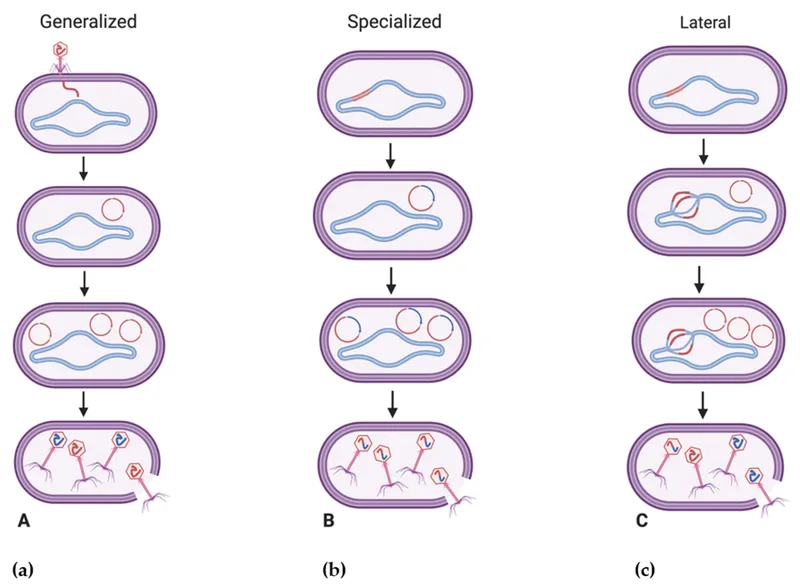
Plasmids and mobile genetic elements US Medical PG Question 7: An investigator studying mechanisms of acquired antibiotic resistance in bacteria conducts a study using isolated strains of Escherichia coli and Staphylococcus aureus. The E. coli strain harbors plasmid pRK212.1, which conveys resistance to kanamycin. The S. aureus strain is susceptible to kanamycin. Both bacterial strains are mixed in a liquid growth medium containing deoxyribonuclease. After incubation for 2 days and subsequent transfer to a solid medium, the S. aureus colonies show no lysis in response to the application of kanamycin. Analysis of chromosomal DNA from the kanamycin-resistant S. aureus strain does not reveal the kanamycin-resistance gene. Which of the following mechanisms is most likely responsible for this finding?
- A. Transformation
- B. Conjugation (Correct Answer)
- C. Transduction
- D. Transposition
- E. Secretion
Plasmids and mobile genetic elements Explanation: ***Conjugation***
- The presence of **deoxyribonuclease (DNase)** in the growth medium inhibits **transformation**, ruling out the uptake of naked DNA. The transfer of the kanamycin resistance gene from a plasmid in *E. coli* to *S. aureus* in the presence of DNase strongly points to **cell-to-cell contact** via conjugation.
- The resistance gene is found on a **plasmid** in *E. coli* and is transferred to *S. aureus*, resulting in kanamycin resistance without integrating into the *S. aureus* chromosome, which is characteristic of conjugative plasmid transfer.
- **Key experimental clue**: DNase destroys free DNA in the medium, so the only way for genetic material to transfer is through **direct cell-to-cell contact**, which is the hallmark of conjugation.
*Transformation*
- This process involves the uptake of **naked DNA** from the environment by a bacterial cell, which would have been prevented by the presence of **deoxyribonuclease** in the medium.
- Transformation typically results in the integration of the foreign DNA into the host cell's **chromosome** or stable maintenance as a plasmid, but DNase would degrade any free DNA before uptake could occur.
*Transduction*
- **Transduction** involves the transfer of genetic material via a **bacteriophage**. The scenario does not describe the presence of any phage particles, nor is there mention of viral vectors.
- The resistance gene originates from a **plasmid** in *E. coli*, and transduction would require a phage capable of infecting both species, which is not mentioned in the experimental design.
*Transposition*
- **Transposition** is the movement of a segment of DNA from one location to another within the **same cell** (e.g., between a plasmid and chromosome). It does not explain the transfer of genetic material **between** two different bacterial cells.
- While a **transposon** might carry the kanamycin resistance gene on the plasmid, transposition itself is not the mechanism for **inter-species transfer** observed in this experiment.
*Secretion*
- **Secretion** refers to the active release of molecules (proteins, enzymes, toxins) from a cell. It is not a mechanism for the direct transfer of **genetic material** (like a plasmid or gene) from one bacterium to another.
- Genetic material is transferred through conjugation, transformation, or transduction, not by secretion pathways.
Plasmids and mobile genetic elements US Medical PG Question 8: A group of microbiological investigators is studying bacterial DNA replication in E. coli colonies. While the cells are actively proliferating, the investigators stop the bacterial cell cycle during S phase and isolate an enzyme involved in DNA replication. An assay of the enzyme's exonuclease activity determines that it is active on both intact and demethylated thymine nucleotides. Which of the following enzymes have the investigators most likely isolated?
- A. DNA ligase
- B. Telomerase
- C. Primase
- D. DNA topoisomerase
- E. DNA polymerase I (Correct Answer)
Plasmids and mobile genetic elements Explanation: ***DNA polymerase I***
- **DNA polymerase I** possesses **5' to 3' exonuclease activity**, which is crucial for removing **RNA primers** (intact nucleotides) laid down by primase during DNA replication.
- This 5' to 3' exonuclease activity also allows it to excise damaged DNA, including DNA containing **demethylated thymine nucleotides**.
- It also has 3' to 5' exonuclease activity for proofreading.
- **Key distinction:** While DNA polymerase III (the main replicative enzyme) only has 3' to 5' exonuclease activity, DNA polymerase I has **both** 3' to 5' and 5' to 3' exonuclease activities, making it essential for primer removal and DNA repair.
*DNA ligase*
- **DNA ligase** functions to form a **phosphodiester bond** between adjacent nucleotides to seal nicks in the DNA backbone, but it does not have exonuclease activity.
- Its primary role is in joining Okazaki fragments and repairing single-strand breaks.
*Telomerase*
- **Telomerase** is a specialized reverse transcriptase that extends the telomeres at the ends of eukaryotic chromosomes, but is not present in prokaryotes like *E. coli*.
- It uses an RNA template to synthesize DNA, and it lacks exonuclease activity.
*Primase*
- **Primase** is an RNA polymerase that synthesizes short **RNA primers** on the DNA template, providing a starting point for DNA synthesis.
- It is involved in synthesizing primers, not in removing or excising nucleotides, and has no exonuclease activity.
*DNA topoisomerase*
- **DNA topoisomerases** relieve supercoiling in DNA during replication and transcription by cutting and rejoining DNA strands.
- While they act on DNA, their function is to manage topological stress, and they do not exhibit exonuclease activity on nucleotides.
Plasmids and mobile genetic elements US Medical PG Question 9: A 29-year-old pregnant woman with no prior antibiotic exposure presents with gonorrhea. Culture of Neisseria gonorrhoeae shows resistance to penicillin, tetracycline, and fluoroquinolones. Genetic testing reveals she has a strain with chromosomal mutations in penA (mosaic allele), mtrR promoter, and gyrA. She reports her partner recently returned from Southeast Asia. Apply epidemiologic and resistance mechanism knowledge to determine the most appropriate management and public health action.
- A. Treat with ceftriaxone alone and report to local health department
- B. Treat with dual therapy (ceftriaxone plus azithromycin) and initiate partner notification with travel history documentation (Correct Answer)
- C. Perform cephalosporin susceptibility testing before treatment initiation
- D. Treat with azithromycin monotherapy due to pregnancy
- E. Initiate spectinomycin therapy and routine partner notification only
Plasmids and mobile genetic elements Explanation: ***Treat with dual therapy (ceftriaxone plus azithromycin) and initiate partner notification with travel history documentation***
- The presence of the **mosaic penA allele** and **mtrR promoter mutations** signifies significant resistance potential; **dual therapy** with ceftriaxone and azithromycin remains critical for ensuring cure and slowing further resistance in highly resistant strains.
- Given the partner's travel to **Southeast Asia**, a region known for emerging **extensively drug-resistant (XDR)** gonorrhea, documenting travel and notification is vital for public health **surveillance**.
*Treat with ceftriaxone alone and report to local health department*
- While ceftriaxone is the backbone of treatment, using **monotherapy** for a strain already exhibiting multiple chromosomal resistance mutations (mosaic penA) increases the risk of selecting for **cephalosporin resistance**.
- This approach is less robust than dual therapy in the context of suspected **imported resistant strains** from high-risk geographic regions.
*Perform cephalosporin susceptibility testing before treatment initiation*
- Delaying treatment for **Neisseria gonorrhoeae** waiting for susceptibility results is inappropriate as it allows for ongoing **transmission** and potential progression to **pelvic inflammatory disease**.
- Clinical guidelines recommend **empiric treatment** based on local prevalence and travel history while simultaneously performing surveillance cultures.
*Treat with azithromycin monotherapy due to pregnancy*
- **Azithromycin monotherapy** is contraindicated for gonorrhea treatment because it has a low barrier to resistance and would fail to cover chromosomal mutations affecting **efflux pumps (mtrR)**.
- While both drugs are safe in **pregnancy**, azithromycin must be used in **combination** with ceftriaxone to prevent treatment failure.
*Initiate spectinomycin therapy and routine partner notification only*
- **Spectinomycin** is a second-line agent that is currently not readily available in the United States and has poor efficacy for **pharyngeal infections**.
- Focusing only on routine notification ignores the critical **epidemiologic significance** of the Southeast Asian travel history associated with highly resistant strains.
Plasmids and mobile genetic elements US Medical PG Question 10: A 67-year-old woman with persistent Enterococcus faecium bacteremia despite appropriate vancomycin therapy undergoes repeat culture. The isolate now shows vancomycin MIC of 128 μg/mL (previously 2 μg/mL). PCR testing reveals the presence of vanA gene cluster. Hospital epidemiology traces potential sources. What is the most likely mechanism by which this organism acquired high-level vancomycin resistance?
- A. Spontaneous chromosomal mutation during therapy
- B. Transposon-mediated transfer from vancomycin-resistant enterococci (Correct Answer)
- C. Transformation with DNA from lysed resistant bacteria
- D. Increased vancomycin efflux pump expression
- E. Alteration in cell wall synthesis without genetic acquisition
Plasmids and mobile genetic elements Explanation: ***Transposon-mediated transfer from vancomycin-resistant enterococci***
- High-level vancomycin resistance in Enterococcus is primarily mediated by the **vanA gene cluster**, which is carried on the **Tn1546 transposon** and spread via **conjugation**.
- This mechanism involves the replacement of the terminal **D-Ala-D-Ala** of peptidoglycan precursors with **D-Ala-D-Lac**, resulting in a 1000-fold decrease in vancomycin binding affinity.
*Spontaneous chromosomal mutation during therapy*
- While mutations can cause resistance to some antibiotics (like rifampin), **high-level vancomycin resistance** in enterococci is not caused by single-point mutations.
- A sudden jump in MIC from 2 to 128 μg/mL is characteristic of **horizontal gene transfer** rather than the gradual accumulation of chromosomal mutations.
*Transformation with DNA from lysed resistant bacteria*
- **Transformation** (uptake of naked DNA) is less common in enterococci compared to **conjugation** for the transfer of large, complex gene clusters like **vanA**.
- The epidemiological tracing implied in the scenario is classic for the spread of **plasmids** and **transposons** between colonized patients in a hospital setting.
*Increased vancomycin efflux pump expression*
- **Efflux pumps** are significant for resistance against drugs like tetracyclines or fluoroquinolones, but they are not the mechanism for **vancomycin resistance**.
- Vancomycin is a large **glycopeptide molecule**; resistance is achieved through **structural modification** of its target (cell wall precursors) rather than active expulsion.
*Alteration in cell wall synthesis without genetic acquisition*
- Vancomycin-intermediate S. aureus (**VISA**) involves cell wall thickening without new gene acquisition, but this results in a **smaller, gradual MIC increase**.
- The detection of the **vanA gene** by PCR confirms that the resistance is due to **acquired genetic material** rather than a purely metabolic or adaptive physiological change.
More Plasmids and mobile genetic elements US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.