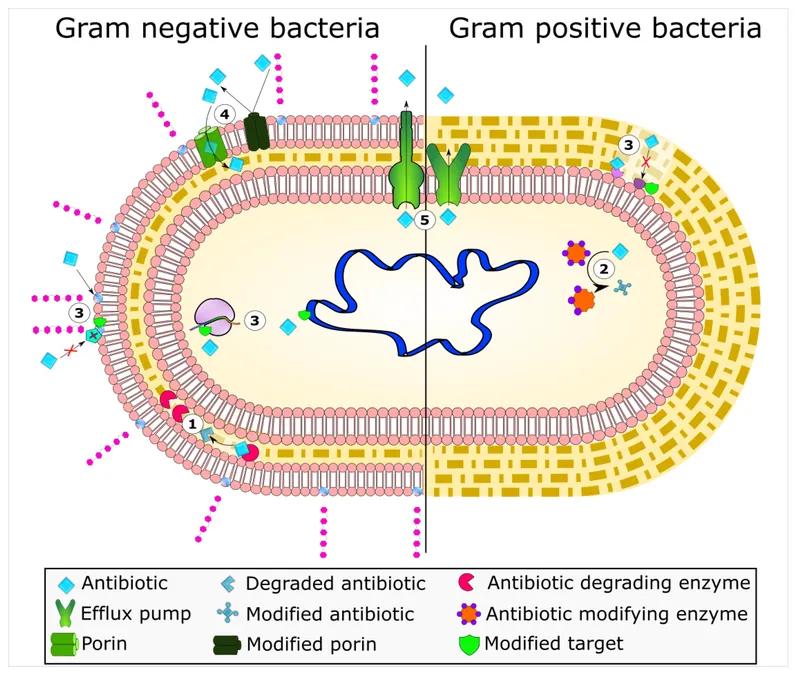
Evolution of antimicrobial resistance US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Evolution of antimicrobial resistance. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Evolution of antimicrobial resistance US Medical PG Question 1: If the genetic material were isolated and injected into the cytoplasm of a human cell, which of the following would produce viable, infectious virions?
- A. Lassa fever virus
- B. Rabies virus
- C. Rhinovirus (Correct Answer)
- D. Mumps virus
- E. Influenza virus
Evolution of antimicrobial resistance Explanation: ***Rhinovirus***
- **Rhinovirus** is a **positive-sense single-stranded RNA virus**. Its genetic material can directly serve as mRNA in the host cell cytoplasm, leading to immediate protein synthesis and viral replication without needing DNA intermediates or a nuclear phase.
- This direct translation allows for the production of viable, infectious virions upon cytoplasmic injection of the genetic material.
*Lassa fever virus*
- **Lassa fever virus** is an **ambisense RNA virus** and requires an RNA-dependent RNA polymerase (RdRp) to transcribe its genome into mRNA.
- This RdRp is packaged within the virion, meaning the injected genetic material alone is not sufficient to initiate replication without the viral proteins.
*Rabies virus*
- **Rabies virus** is a **negative-sense single-stranded RNA virus**. Its genome cannot directly act as mRNA.
- It requires a virion-associated **RNA-dependent RNA polymerase (RdRp)** to transcribe its negative-sense RNA into positive-sense mRNA, which is essential for protein synthesis.
*Mumps virus*
- **Mumps virus** is a **negative-sense single-stranded RNA virus** and, like rabies virus, cannot directly translate its genome into proteins.
- It also requires its own **virion-associated RNA-dependent RNA polymerase** to synthesize mRNA from its negative-sense genome.
*Influenza virus*
- **Influenza virus** is a **negative-sense segmented RNA virus**. Its replication cycle involves the nucleus, where its RNA genome is transcribed into mRNA.
- This process requires the viral **RNA-dependent RNA polymerase**, which is brought into the cell by the virion, and interaction with host nuclear machinery.
Evolution of antimicrobial resistance US Medical PG Question 2: Six days after undergoing an elective hip replacement surgery, a 79-year-old man develops dysuria, flank pain, and fever. His temperature is 38.5°C (101.3°F). Examination shows marked tenderness in the right costovertebral area. Treatment with an antibiotic is begun, but his symptoms do not improve. Further evaluation shows that the causal organism produces an enzyme that inactivates the antibiotic via phosphorylation. An agent from which of the following classes of antibiotics was most likely administered?
- A. Macrolides
- B. Tetracyclines
- C. Aminoglycosides (Correct Answer)
- D. Glycopeptides
- E. Fluoroquinolones
Evolution of antimicrobial resistance Explanation: ***Aminoglycosides***
- **Aminoglycosides** are commonly inactivated by bacterial enzymes through **phosphorylation**, acetylation, or adenylation, leading to resistance.
- The patient's lack of improvement despite antibiotic treatment and the mechanism of inactivation point towards this class of antibiotics.
*Macrolides*
- **Macrolide resistance** typically involves mechanisms such as modification of the ribosomal binding site (e.g., methylation), drug efflux pumps, or enzymatic inactivation by esterases, not phosphorylation.
- While macrolides can treat various infections, their inactivation mechanism is different from what is described.
*Tetracyclines*
- **Tetracycline resistance** is primarily mediated by bacterial efflux pumps that actively transport the antibiotic out of the cell, or by ribosomal protection proteins that interfere with drug binding.
- **Enzymatic inactivation via phosphorylation** is not a characteristic resistance mechanism for tetracyclines.
*Glycopeptides*
- **Glycopeptide resistance**, particularly to vancomycin, is mainly associated with alterations in the cell wall precursor target (e.g., D-Ala-D-Lac modification), which prevents the antibiotic from binding.
- This mechanism is distinct from enzymatic phosphorylation of the antibiotic molecule itself.
*Fluoroquinolones*
- **Fluoroquinolone resistance** primarily arises from mutations in the genes encoding bacterial DNA gyrase and topoisomerase IV, or via efflux pumps.
- There is no significant mechanism of resistance involving direct enzymatic phosphorylation of fluoroquinolone drugs.
Evolution of antimicrobial resistance US Medical PG Question 3: An 18-year old college freshman presents to his university clinic because he has not been feeling well for the past two weeks. He has had a persistent headache, occasional cough, and chills without rigors. The patient’s vital signs are normal and physical exam is unremarkable. His radiograph shows patchy interstitial lung infiltrates and he is diagnosed with atypical pneumonia. The patient is prescribed azithromycin and takes his medication as instructed. Despite adherence to his drug regimen, he returns to the clinic one week later because his symptoms have not improved. The organism responsible for this infection is likely resistant to azithromycin through which mechanism?
- A. Mutation in topoisomerase II
- B. Methylation of ribosomal binding site
- C. Presence of a beta-lactamase
- D. Decreased binding to RNA polymerase
- E. Insertion of drug efflux pumps (Correct Answer)
Evolution of antimicrobial resistance Explanation: ***Insertion of drug efflux pumps***
- **Azithromycin** is a macrolide antibiotic that inhibits bacterial protein synthesis by binding to the **50S ribosomal subunit**.
- In **Mycoplasma pneumoniae** (the most common cause of atypical pneumonia in young adults), the **most common** mechanism of macrolide resistance is through **efflux pumps**, particularly the **mef genes**.
- These efflux pumps actively transport macrolides out of the bacterial cell, reducing intracellular drug concentration and conferring resistance.
- This mechanism is responsible for the majority of macrolide-resistant *M. pneumoniae* isolates worldwide.
*Methylation of ribosomal binding site*
- **Methylation** of the ribosomal binding site (specifically the **23S rRNA** via erm genes) does prevent azithromycin from binding effectively.
- While this is a valid macrolide resistance mechanism seen in organisms like *Streptococcus pneumoniae* and *Streptococcus pyogenes*, it is **less common** in *Mycoplasma pneumoniae*.
- Efflux pumps (mef) are the predominant mechanism in *M. pneumoniae* resistant strains.
*Mutation in topoisomerase II*
- **Topoisomerase II** (DNA gyrase) is the target of **fluoroquinolone antibiotics**, not macrolides.
- Mutations in this enzyme lead to resistance against fluoroquinolones, such as **ciprofloxacin**.
*Presence of a beta-lactamase*
- **Beta-lactamase enzymes** inactivate **beta-lactam antibiotics** (e.g., penicillin, cephalosporins) by hydrolyzing their beta-lactam ring.
- Additionally, *Mycoplasma pneumoniae* **lacks a cell wall**, making it inherently resistant to all beta-lactam antibiotics regardless of beta-lactamase production.
*Decreased binding to RNA polymerase*
- **RNA polymerase** is the target for antibiotics like **rifampin**, which inhibits bacterial transcription.
- Decreased binding to RNA polymerase would lead to rifampin resistance, not azithromycin resistance.
Evolution of antimicrobial resistance US Medical PG Question 4: A 64-year-old female with type 2 diabetes mellitus comes to the physician because of a 1-week history of painful red swelling on her left thigh. Examination shows a 3- x 4-cm, tender, fluctuant mass. Incision and drainage of the abscess are performed. Culture of the abscess fluid grows gram-positive, coagulase-positive cocci that are resistant to oxacillin. Which of the following best describes the mechanism of resistance of the causal organism to oxacillin?
- A. Degradation of the antibiotic
- B. Decreased uptake of the antibiotic
- C. Decreased activation of the antibiotic
- D. Altered target of the antibiotic (Correct Answer)
- E. Acetylation of the antibiotic
Evolution of antimicrobial resistance Explanation: ***Altered target of the antibiotic***
- The organism described (gram-positive, coagulase-positive cocci, oxacillin-resistant) is **methicillin-resistant *Staphylococcus aureus* (MRSA)**.
- MRSA achieves oxacillin (and other beta-lactam) resistance by acquiring the ***mecA* gene**, which encodes for a **modified penicillin-binding protein (PBP2a)** with reduced affinity for beta-lactam antibiotics.
*Degradation of the antibiotic*
- This mechanism, primarily through the production of **beta-lactamase enzymes**, can degrade beta-lactam antibiotics.
- While *Staphylococcus aureus* can produce beta-lactamases, oxacillin (a **penicillinase-resistant penicillin**) is specifically engineered to be stable against these enzymes.
*Decreased uptake of the antibiotic*
- Reduced permeability of the bacterial cell wall can lead to decreased uptake, a mechanism more commonly associated with **gram-negative bacteria** due to their outer membrane.
- This is not the primary mechanism of resistance for MRSA to oxacillin.
*Decreased activation of the antibiotic*
- Some antibiotics are prodrugs that require activation by bacterial enzymes, and resistance can arise from mutations affecting this activation.
- Oxacillin is active in its administered form and does not require bacterial activation.
*Acetylation of the antibiotic*
- **Enzymatic modification**, such as acetylation, adenylylation, or phosphorylation, is a common mechanism of resistance, particularly against **aminoglycoside antibiotics**.
- This specific mechanism is not responsible for oxacillin resistance in MRSA.
Evolution of antimicrobial resistance US Medical PG Question 5: A 32-year-old man presents to an outpatient clinic for tuberculosis prophylaxis before leaving for a trip to Asia, where tuberculosis is endemic. The Mantoux test is positive, but the chest X-ray and AFB sputum culture are negative. He was started on isoniazid. What is the most likely mechanism of resistance to isoniazid?
- A. Methylation of the RNA binding site
- B. Plasmid-mediated resistance
- C. Reduction of drug binding to RNA polymerase
- D. Increased efflux from the cell
- E. Mutations in katG (Correct Answer)
Evolution of antimicrobial resistance Explanation: ***Mutations in katG***
- The **katG gene** encodes **catalase-peroxidase**, an enzyme essential for activating isoniazid into its active form within *Mycobacterium tuberculosis*.
- Mutations in *katG* prevent the activation of isoniazid, thereby conferring **resistance**.
*Methylation of the RNA binding site*
- This mechanism is primarily associated with **aminoglycoside resistance**, where methylation of ribosomal RNA prevents antibiotic binding.
- It is not a known mechanism for resistance to **isoniazid**.
*Plasmid-mediated resistance*
- While common in many bacteria for antibiotic resistance, **plasmid-mediated resistance** is rare for **first-line anti-tuberculosis drugs** like isoniazid in *Mycobacterium tuberculosis*.
- Most *M. tuberculosis* resistance mechanisms involve **chromosomal mutations**.
*Reduction of drug binding to RNA polymerase*
- This mechanism is typically associated with resistance to **rifamycins** (e.g., rifampin), which target the **bacterial RNA polymerase**.
- Isoniazid's mechanism of action involves **mycolic acid synthesis inhibition**, not RNA polymerase binding.
*Increased efflux from the cell*
- While efflux pumps contribute to antibiotic resistance in many bacteria, they are less commonly the primary mechanism for high-level **isoniazid resistance** in *M. tuberculosis*.
- Resistance is predominantly linked to target modification or enzyme deficits, like those involving **katG**.
Evolution of antimicrobial resistance US Medical PG Question 6: An investigator is studying bacterial toxins in a nonpathogenic bacterial monoculture that has been inoculated with specific bacteriophages. These phages were previously cultured in a toxin-producing bacterial culture. After inoculation, a new toxin is isolated from the culture. Genetic sequencing shows that the bacteria have incorporated viral genetic information, including the gene for this toxin, into their genome. The described process is most likely responsible for acquired pathogenicity in which of the following bacteria?
- A. Staphylococcus aureus
- B. Haemophilus influenzae
- C. Neisseria meningitidis
- D. Streptococcus pneumoniae
- E. Corynebacterium diphtheriae (Correct Answer)
Evolution of antimicrobial resistance Explanation: ***Corynebacterium diphtheriae***
- The process described, where a bacterium acquires new genetic information (e.g., a toxin gene) from a bacteriophage, is called **lysogenic conversion** or **phage conversion**. *Corynebacterium diphtheriae* is the **classic example** of this mechanism, acquiring its toxigenicity through phage-mediated transfer of the **diphtheria toxin gene (tox gene)** via bacteriophage β.
- The diphtheria toxin is an **AB toxin** that ADP-ribosylates and thereby inactivates **elongation factor 2 (EF-2)**, inhibiting host cell protein synthesis and leading to the characteristic symptoms of diphtheria.
- This is the **prototypical and most clinically significant example** of lysogenic conversion in medical microbiology.
*Staphylococcus aureus*
- While *Staphylococcus aureus* can acquire some virulence factors via bacteriophages (e.g., **Panton-Valentine leukocidin**, some enterotoxins), many of its toxins are encoded on **mobile genetic elements** such as plasmids, pathogenicity islands, or chromosomal genes.
- However, *S. aureus* is **not the classic example** of lysogenic conversion described in this scenario. *C. diphtheriae* better exemplifies the acquisition of a major toxin exclusively through phage conversion.
*Haemophilus influenzae*
- *Haemophilus influenzae* primarily causes disease through its **polysaccharide capsule** (especially type b) and is a common cause of respiratory infections and meningitis.
- Its major virulence factors are typically chromosomally encoded or acquired through **transformation** (uptake of naked DNA), not through phage conversion for a primary toxin.
*Neisseria meningitidis*
- *Neisseria meningitidis* causes meningococcal disease, primarily due to its **polysaccharide capsule** and **endotoxin (LPS)**.
- While genetic exchange can occur, the acquisition of a major toxin gene by phage conversion as described is not a primary mechanism for its key virulence factors.
*Streptococcus pneumoniae*
- *Streptococcus pneumoniae* is a leading cause of pneumonia, meningitis, and otitis media, with its main virulence factor being its **polysaccharide capsule**.
- It primarily acquires genetic material through **transformation** (competence-mediated uptake of naked DNA), which contributes to antibiotic resistance and capsule types, but lysogenic conversion with toxin acquisition is not typical for its major virulence factors.
Evolution of antimicrobial resistance US Medical PG Question 7: A 61-year-old woman who recently emigrated from India comes to the physician because of a 2-month history of fever, fatigue, night sweats, and a productive cough. She has had a 5-kg (11-lb) weight loss during this period. She has a history of type 2 diabetes mellitus and poorly controlled asthma. She has had multiple asthma exacerbations in the past year that were treated with glucocorticoids. An x-ray of the chest shows a cavitary lesion of the posterior apical segment of the left upper lobe with consolidation of the surrounding parenchyma. The pathogen identified on sputum culture is found to be resistant to multiple drugs, including streptomycin. Which of the following mechanisms is most likely involved in bacterial resistance to this drug?
- A. Alteration in the sequence of gyrA genes
- B. Upregulation of arabinosyl transferase production
- C. Upregulation of mycolic acid synthesis
- D. Alteration in 30S ribosomal subunit (Correct Answer)
- E. Inhibition of bacterial synthesis of RNA
Evolution of antimicrobial resistance Explanation: ***Alteration in 30S ribosomal subunit***
- Streptomycin is an **aminoglycoside antibiotic** that acts by binding to the **16S rRNA of the 30S ribosomal subunit**, which interferes with bacterial protein synthesis.
- **Resistance to streptomycin** most commonly arises from mutations in the genes encoding ribosomal proteins (e.g., *rpsL*) or the 16S rRNA that alter the drug's binding site on the 30S ribosomal subunit, preventing its inhibitory effect.
*Alteration in the sequence of gyrA genes*
- Mutations in the *gyrA* gene typically confer resistance to **fluoroquinolone antibiotics**, such as ciprofloxacin and levofloxacin.
- Fluoroquinolones target **DNA gyrase (topoisomerase II)**, which is encoded by *gyrA*, not the ribosomes.
*Upregulation of arabinosyl transferase production*
- **Arabinogalactan**, a major component of the mycobacterial cell wall, is synthesized by **arabinosyl transferases** (e.g., EmbB).
- Resistance to **ethambutol** is often associated with mutations or upregulation of these enzymes, leading to increased synthesis of the arabinogalactan layer.
*Upregulation of mycolic acid synthesis*
- **Mycolic acid** is a crucial component of the mycobacterial cell wall, and its synthesis is inhibited by drugs like **isoniazid**.
- Upregulation of mycolic acid synthesis or mutations in genes related to its production (e.g., *kasA*) can lead to **isoniazid resistance**, but not directly to streptomycin resistance.
*Inhibition of bacterial synthesis of RNA*
- **Rifampin** is an antibiotic that inhibits bacterial RNA synthesis by binding to the **DNA-dependent RNA polymerase**.
- While resistance to rifampin often involves mutations in the *rpoB* gene, this mechanism is specific to rifampin and not streptomycin.
Evolution of antimicrobial resistance US Medical PG Question 8: A 55-year-old man who recently immigrated to the United States from Azerbaijan comes to the physician because of a 6-week history of recurrent fever, progressive cough with bloody streaks, fatigue, and a 3.6-kg (8-lb) weight loss. He has poorly-controlled type 2 diabetes mellitus treated with insulin. An x-ray of the chest shows a cavitary lesion of the posterior apical segment of the right upper lobe with consolidation of the surrounding parenchyma. He is started on a treatment regimen with a combination of drugs. A culture of the sputum identifies a causal pathogen that is resistant to a drug that alters the metabolism of pyridoxine. Which of the following is the most likely mechanism of resistance to this drug?
- A. Increased production of arabinosyl transferase
- B. Mutation in genes encoding RNA polymerase
- C. Changed amino acid composition of DNA gyrase
- D. Impaired conversion to pyrazinoic acid
- E. Decreased production of catalase-peroxidase (Correct Answer)
Evolution of antimicrobial resistance Explanation: ***Decreased production of catalase-peroxidase***
- The drug described as altering the metabolism of pyridoxine is **isoniazid**, which requires activation by the catalase-peroxidase enzyme **KatG** in *Mycobacterium tuberculosis*.
- Resistance to isoniazid often develops through **mutations or deletions in the *katG* gene**, leading to decreased or absent catalase-peroxidase activity and thus impaired activation of the drug.
*Increased production of arabinosyl transferase*
- This mechanism is associated with resistance to **ethambutol**, which inhibits **arabinosyl transferase**, an enzyme crucial for cell wall synthesis.
- Increased production of this enzyme would counteract the drug's effect, but it is not relevant to the drug that alters pyridoxine metabolism (isoniazid).
*Mutation in genes encoding RNA polymerase*
- This mechanism is responsible for resistance to **rifampin**, which targets the **bacterial DNA-dependent RNA polymerase**.
- Mutations in the *rpoB* gene prevent rifampin from binding, but this drug does not alter pyridoxine metabolism.
*Changed amino acid composition of DNA gyrase*
- This mechanism is characteristic of resistance to **fluoroquinolones** (e.g., ciprofloxacin, levofloxacin), which inhibit **DNA gyrase** and topoisomerase IV.
- Alterations in the drug-binding sites of these enzymes reduce the efficacy of fluoroquinolones, a different class of antibiotics from isoniazid.
*Impaired conversion to pyrazinoic acid*
- This is the primary mechanism of resistance to **pyrazinamide**, which is a prodrug that needs to be converted to its active form, **pyrazinoic acid**, by the bacterial enzyme **pyrazinamidase (PncA)**.
- Resistance typically involves mutations in the *pncA* gene, but pyrazinamide is not the drug that alters pyridoxine metabolism.
Evolution of antimicrobial resistance US Medical PG Question 9: A 29-year-old pregnant woman with no prior antibiotic exposure presents with gonorrhea. Culture of Neisseria gonorrhoeae shows resistance to penicillin, tetracycline, and fluoroquinolones. Genetic testing reveals she has a strain with chromosomal mutations in penA (mosaic allele), mtrR promoter, and gyrA. She reports her partner recently returned from Southeast Asia. Apply epidemiologic and resistance mechanism knowledge to determine the most appropriate management and public health action.
- A. Treat with ceftriaxone alone and report to local health department
- B. Treat with dual therapy (ceftriaxone plus azithromycin) and initiate partner notification with travel history documentation (Correct Answer)
- C. Perform cephalosporin susceptibility testing before treatment initiation
- D. Treat with azithromycin monotherapy due to pregnancy
- E. Initiate spectinomycin therapy and routine partner notification only
Evolution of antimicrobial resistance Explanation: ***Treat with dual therapy (ceftriaxone plus azithromycin) and initiate partner notification with travel history documentation***
- The presence of the **mosaic penA allele** and **mtrR promoter mutations** signifies significant resistance potential; **dual therapy** with ceftriaxone and azithromycin remains critical for ensuring cure and slowing further resistance in highly resistant strains.
- Given the partner's travel to **Southeast Asia**, a region known for emerging **extensively drug-resistant (XDR)** gonorrhea, documenting travel and notification is vital for public health **surveillance**.
*Treat with ceftriaxone alone and report to local health department*
- While ceftriaxone is the backbone of treatment, using **monotherapy** for a strain already exhibiting multiple chromosomal resistance mutations (mosaic penA) increases the risk of selecting for **cephalosporin resistance**.
- This approach is less robust than dual therapy in the context of suspected **imported resistant strains** from high-risk geographic regions.
*Perform cephalosporin susceptibility testing before treatment initiation*
- Delaying treatment for **Neisseria gonorrhoeae** waiting for susceptibility results is inappropriate as it allows for ongoing **transmission** and potential progression to **pelvic inflammatory disease**.
- Clinical guidelines recommend **empiric treatment** based on local prevalence and travel history while simultaneously performing surveillance cultures.
*Treat with azithromycin monotherapy due to pregnancy*
- **Azithromycin monotherapy** is contraindicated for gonorrhea treatment because it has a low barrier to resistance and would fail to cover chromosomal mutations affecting **efflux pumps (mtrR)**.
- While both drugs are safe in **pregnancy**, azithromycin must be used in **combination** with ceftriaxone to prevent treatment failure.
*Initiate spectinomycin therapy and routine partner notification only*
- **Spectinomycin** is a second-line agent that is currently not readily available in the United States and has poor efficacy for **pharyngeal infections**.
- Focusing only on routine notification ignores the critical **epidemiologic significance** of the Southeast Asian travel history associated with highly resistant strains.
Evolution of antimicrobial resistance US Medical PG Question 10: A 67-year-old woman with persistent Enterococcus faecium bacteremia despite appropriate vancomycin therapy undergoes repeat culture. The isolate now shows vancomycin MIC of 128 μg/mL (previously 2 μg/mL). PCR testing reveals the presence of vanA gene cluster. Hospital epidemiology traces potential sources. What is the most likely mechanism by which this organism acquired high-level vancomycin resistance?
- A. Spontaneous chromosomal mutation during therapy
- B. Transposon-mediated transfer from vancomycin-resistant enterococci (Correct Answer)
- C. Transformation with DNA from lysed resistant bacteria
- D. Increased vancomycin efflux pump expression
- E. Alteration in cell wall synthesis without genetic acquisition
Evolution of antimicrobial resistance Explanation: ***Transposon-mediated transfer from vancomycin-resistant enterococci***
- High-level vancomycin resistance in Enterococcus is primarily mediated by the **vanA gene cluster**, which is carried on the **Tn1546 transposon** and spread via **conjugation**.
- This mechanism involves the replacement of the terminal **D-Ala-D-Ala** of peptidoglycan precursors with **D-Ala-D-Lac**, resulting in a 1000-fold decrease in vancomycin binding affinity.
*Spontaneous chromosomal mutation during therapy*
- While mutations can cause resistance to some antibiotics (like rifampin), **high-level vancomycin resistance** in enterococci is not caused by single-point mutations.
- A sudden jump in MIC from 2 to 128 μg/mL is characteristic of **horizontal gene transfer** rather than the gradual accumulation of chromosomal mutations.
*Transformation with DNA from lysed resistant bacteria*
- **Transformation** (uptake of naked DNA) is less common in enterococci compared to **conjugation** for the transfer of large, complex gene clusters like **vanA**.
- The epidemiological tracing implied in the scenario is classic for the spread of **plasmids** and **transposons** between colonized patients in a hospital setting.
*Increased vancomycin efflux pump expression*
- **Efflux pumps** are significant for resistance against drugs like tetracyclines or fluoroquinolones, but they are not the mechanism for **vancomycin resistance**.
- Vancomycin is a large **glycopeptide molecule**; resistance is achieved through **structural modification** of its target (cell wall precursors) rather than active expulsion.
*Alteration in cell wall synthesis without genetic acquisition*
- Vancomycin-intermediate S. aureus (**VISA**) involves cell wall thickening without new gene acquisition, but this results in a **smaller, gradual MIC increase**.
- The detection of the **vanA gene** by PCR confirms that the resistance is due to **acquired genetic material** rather than a purely metabolic or adaptive physiological change.
More Evolution of antimicrobial resistance US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.