Microbial genetics and drug resistance

On this page
🧬 The Genetic Arsenal: Microbial Warfare Strategies
Bacteria don't just survive antibiotics-they share blueprints for resistance like intelligence networks, transforming individual mutations into collective threats that spread through hospitals and communities. You'll discover how microbes weaponize their genetic flexibility through horizontal gene transfer, mobile elements, and plasmids, then trace how these molecular strategies create the multidrug-resistant superbugs that challenge modern medicine. Understanding this genetic warfare reveals both why resistance emerges so rapidly and where we might intervene to preserve our therapeutic arsenal.
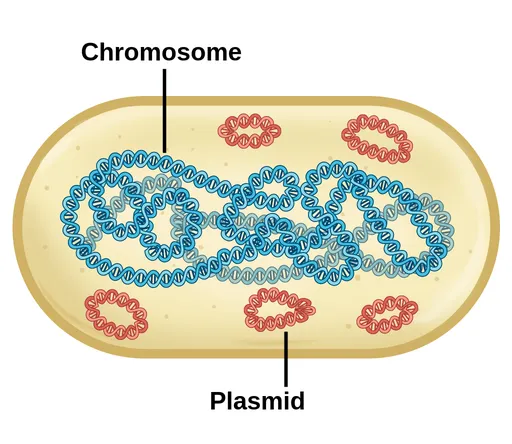
The Bacterial Genetic Command Center
Bacterial genetics operates through multiple interconnected systems that enable rapid adaptation and survival:
-
Chromosomal DNA
- Single circular chromosome (1-10 million base pairs)
- Essential genes for survival and basic functions
- Integration sites for mobile genetic elements
- Insertion sequences (IS elements): 768-5,000 base pairs
- Prophage integration: 20-60 kb typical size
- Chromosomal resistance islands: 10-200 kb clusters
-
Extrachromosomal Elements
- Plasmids: 1.5-400 kb autonomous replicating DNA
- Episomes: can integrate into chromosome or exist independently
- Megaplasmids: >100 kb carrying multiple resistance genes
- R plasmids: resistance gene clusters
- Virulence plasmids: pathogenicity factors
- Metabolic plasmids: specialized enzyme systems
📌 Remember: CHROME - Chromosome Holds Resistance Origins, Mobile Elements spread them
Resistance Gene Architecture
The organization of resistance genes follows predictable patterns that enable rapid dissemination:
| Element Type | Size Range | Copy Number | Mobility | Clinical Impact |
|---|---|---|---|---|
| Insertion Sequences | 0.7-5 kb | 1-50 copies | High | Activates resistance |
| Transposons | 2-150 kb | 1-10 copies | Moderate | Transfers clusters |
| Integrons | 0.5-200 kb | 1-5 copies | Low | Captures cassettes |
| Plasmids | 1.5-400 kb | 1-20 copies | High | Spreads resistance |
| ICEs | 18-500 kb | 1 copy | Moderate | Transfers virulence |
Mobile Genetic Element Hierarchy
💡 Master This: Mobile genetic elements create a hierarchical resistance network - plasmids carry transposons, transposons carry resistance genes, and integrons capture gene cassettes, enabling exponential resistance spread.
The genetic foundation reveals how bacteria transform from susceptible organisms into multidrug-resistant superbugs through coordinated molecular mechanisms. Understanding horizontal gene transfer mechanisms unlocks the logic behind resistance emergence patterns.
🧬 The Genetic Arsenal: Microbial Warfare Strategies
⚡ Horizontal Gene Highway: The Resistance Superhighway

Conjugation: The Bacterial Internet
Conjugation functions as the most clinically significant mechanism for resistance dissemination:
-
Pilus Formation and Contact
- F pilus extends 1-20 μm from donor cell
- Contact establishment within 2-5 minutes
- Pilus retraction brings cells within 10-50 nm
- Mating bridge formation: 30-60 seconds
- DNA transfer initiation: 1-2 minutes
- Complete plasmid transfer: 30-100 minutes
-
Transfer Efficiency Factors
- Temperature optimum: 25-37°C (90% efficiency)
- pH range: 6.5-8.0 (optimal 7.2-7.4)
- Nutrient availability increases transfer 5-10 fold
- Rich media: 10⁻² to 10⁻¹ transfer frequency
- Minimal media: 10⁻⁴ to 10⁻³ transfer frequency
- Biofilm conditions: 100-1000x enhanced transfer
📌 Remember: BRIDGE - Bacterial Resistance Increases During Gene Exchange
Transformation: Environmental DNA Scavenging
Natural transformation enables bacteria to acquire resistance from environmental DNA:
| Bacterial Species | Competence Type | Efficiency | Clinical Relevance |
|---|---|---|---|
| Streptococcus pneumoniae | Natural | 10⁻² to 10⁻¹ | PBP mutations |
| Haemophilus influenzae | Natural | 10⁻³ to 10⁻² | β-lactamase genes |
| Neisseria gonorrhoeae | Natural | 10⁻³ to 10⁻² | Multiple resistance |
| Acinetobacter baumannii | Natural | 10⁻⁴ to 10⁻³ | Carbapenemase genes |
| Pseudomonas aeruginosa | Induced | 10⁻⁵ to 10⁻⁴ | Efflux pumps |
Transduction: Viral-Mediated Gene Transfer
Bacteriophages serve as vectors for resistance gene transfer between bacterial cells:
-
Generalized Transduction
- Occurs during lytic cycle: 1 in 1,000 phage particles
- Random DNA packaging: 0.5-2% of bacterial genome
- Transfer frequency: 10⁻⁴ to 10⁻⁵ per phage infection
- P1 phage in E. coli: 2% packaging error rate
- Mu phage: 5% transducing particle frequency
- Lambda phage: 0.1% generalized transduction
-
Specialized Transduction
- Prophage excision errors: 1 in 10,000 events
- Adjacent gene transfer: 10-50 kb typical size
- Site-specific integration: 99% accuracy at attP sites
- Lambda phage: gal and bio operons
- P1 phage: random integration sites
- Mu phage: 50+ integration sites per genome
💡 Master This: Horizontal gene transfer creates resistance networks where a single resistant bacterium can transfer genes to 10³-10⁶ recipient cells within 24-48 hours under optimal conditions.
Understanding transfer mechanisms reveals how resistance genes move between bacterial populations. Mobile genetic elements provide the vehicles for this genetic cargo transport system.
⚡ Horizontal Gene Highway: The Resistance Superhighway
🚛 Mobile Genetic Cargo: The Resistance Transport Fleet
Transposons: The Jumping Gene Specialists
Transposons represent autonomous genetic units capable of moving between different DNA molecules:
-
Composite Transposons (Class I)
- Structure: IS element + resistance genes + IS element
- Size range: 5-40 kb typical constructs
- Movement frequency: 10⁻⁵ to 10⁻³ per generation
- Tn10 (tetracycline): 10⁻⁴ transposition rate
- Tn5 (kanamycin): 10⁻⁵ transposition rate
- Tn903 (kanamycin): 10⁻⁶ transposition rate
-
Complex Transposons (Class II)
- Single transposase gene with terminal inverted repeats
- Size range: 2-150 kb resistance clusters
- Integration specificity: 5-9 bp target site duplications
- Tn916 (tetracycline): 18 kb, broad host range
- Tn1546 (vancomycin): 10.8 kb, VRE-associated
- Tn4001 (gentamicin): 4.9 kb, staphylococcal
📌 Remember: JUMP - Joining Units Move Plasmids (transposons facilitate plasmid-to-chromosome integration)
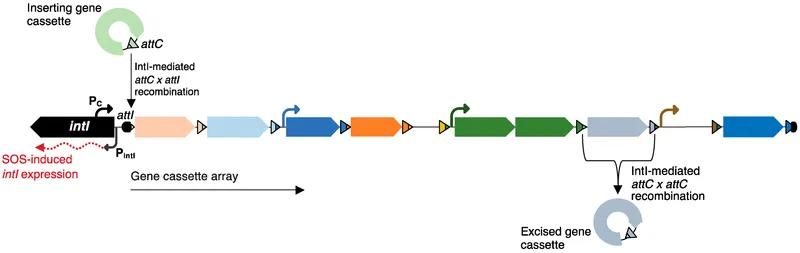
Integrons: The Gene Cassette Collection System
Integrons function as genetic platforms that capture and express gene cassettes:
| Integron Class | IntI Enzyme | Resistance Genes | Clinical Distribution |
|---|---|---|---|
| Class 1 | IntI1 | β-lactamases, aminoglycosides | 95% clinical isolates |
| Class 2 | IntI2 | Trimethoprim, streptomycin | 5% clinical isolates |
| Class 3 | IntI3 | β-lactamases, chloramphenicol | <1% clinical isolates |
| Class 4 | IntI4 | Vibrio-specific resistance | Marine environments |
| Class 5 | IntI5 | Trimethoprim resistance | Emerging clinical |
- attI site recognition: 7 bp core sequence (GTTRRRY)
- Integrase-mediated recombination: 99% site-specific
- Cassette integration time: 15-30 minutes in vitro
- Single cassette integration: 10⁻² to 10⁻¹ efficiency
- Multiple cassette arrays: 60+ genes maximum observed
- Expression gradient: 5'-proximal > distal (10-fold difference)
⭐ Clinical Pearl: Class 1 integrons are found in 95% of multidrug-resistant gram-negative clinical isolates, making them the most successful resistance capture system.
Insertion Sequences: The Genetic Activators
Insertion sequences provide the mobility machinery for composite transposons:
-
IS Element Classification
- IS1 family: 768 bp, DDE transposase motif
- IS3 family: 1,258 bp, two-step transposition
- IS4 family: 1,428 bp, target site specificity
- IS26: 820 bp, broad distribution in Enterobacteriaceae
- IS256: 1,324 bp, staphylococcal resistance activation
- IS6110: 1,355 bp, mycobacterial RFLP marker
-
Resistance Gene Activation Mechanisms
- Promoter insertion: 10-100 fold expression increase
- Gene fusion events: hybrid protein formation
- Regulatory disruption: constitutive expression
- β-lactamase activation: 90% involve IS elements
- Efflux pump upregulation: 5-50 fold increase
- Porin downregulation: 50-90% reduction
💡 Master This: Mobile genetic elements create modular resistance systems where individual components can be mixed, matched, and transferred independently, generating infinite resistance combinations.
Mobile genetic elements provide the transport infrastructure for resistance dissemination. Plasmids serve as the primary vehicles carrying these genetic cargo systems between bacterial cells.
🚛 Mobile Genetic Cargo: The Resistance Transport Fleet
🎯 Plasmid Command Centers: The Resistance Motherships

Plasmid Architecture and Classification
Plasmids exhibit sophisticated organizational structures optimized for resistance gene maintenance and transfer:
-
Size-Based Classification
- Small plasmids: 1.5-10 kb (single resistance gene)
- Medium plasmids: 10-100 kb (multiple resistance clusters)
- Large plasmids: 100-400 kb (resistance + virulence)
- pUC series: 2.7 kb, cloning vectors
- R1 plasmid: 97 kb, multidrug resistance
- pTi plasmids: 200-800 kb, Agrobacterium virulence
-
Functional Modules
- Replication origin (oriV): 200-500 bp essential region
- Partition genes (par): 2-4 genes ensuring inheritance
- Transfer genes (tra): 20-35 genes for conjugation
- Relaxase complex: 3-5 proteins
- Pilus assembly: 12-15 structural proteins
- Transfer regulation: 5-8 regulatory proteins
📌 Remember: PARTS - Plasmids Always Require Transfer Systems (for resistance spread)
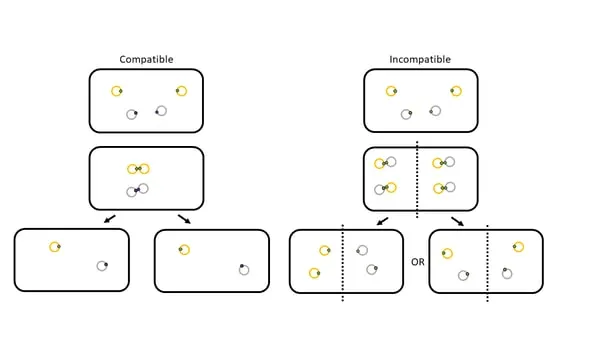
Incompatibility Groups and Host Range
Plasmid incompatibility determines coexistence patterns and resistance gene combinations:
| Inc Group | Host Range | Resistance Profile | Clinical Significance |
|---|---|---|---|
| IncF | Enterobacteriaceae | β-lactamases, quinolones | 40% clinical isolates |
| IncI | Broad gram-negative | Aminoglycosides, sulfonamides | 25% clinical isolates |
| IncN | Very broad | Multiple drug classes | 15% clinical isolates |
| IncP | Extremely broad | Heavy metals, antibiotics | 10% clinical isolates |
| IncW | Broad gram-negative | β-lactamases, tetracycline | 8% clinical isolates |
- Replication control conflicts: same origin cannot coexist
- Partition system interference: unequal segregation
- Surface exclusion: prevents superinfection
- Compatible plasmids: 5-10 per cell maximum
- Incompatible plasmids: mutual exclusion
- Plasmid curing: 10⁻² to 10⁻³ spontaneous loss
⭐ Clinical Pearl: IncF plasmids carry 60-80% of ESBL genes in E. coli, making them the primary vehicles for β-lactam resistance in Enterobacteriaceae.
Resistance Gene Organization Patterns
Plasmids organize resistance genes in predictable clusters that reflect selective pressures:
-
Gene Cluster Architecture
- Resistance islands: 10-50 kb contiguous regions
- Transposon insertions: nested mobile elements
- Integron platforms: gene cassette arrays
- Class 1 integrons: 95% of resistance clusters
- Multiple resistance regions: 2-5 per large plasmid
- Regulatory elements: promoters, terminators
-
Co-resistance Patterns
- β-lactam + aminoglycoside: 85% co-occurrence
- Quinolone + sulfonamide: 70% co-occurrence
- Tetracycline + chloramphenicol: 60% co-occurrence
- Triple resistance: 40% of clinical isolates
- Quadruple resistance: 25% of clinical isolates
- Penta-resistance: 15% of clinical isolates
💡 Master This: Plasmids function as genetic integration platforms that combine individual resistance mechanisms into synergistic resistance networks, creating the multidrug-resistant phenotypes that define modern antimicrobial resistance challenges.
Plasmid-mediated resistance creates the foundation for understanding how individual resistance mechanisms combine into clinical phenotypes. Specific resistance mechanisms reveal the molecular details of antimicrobial failure.
🎯 Plasmid Command Centers: The Resistance Motherships
🛡️ Molecular Defense Arsenal: The Resistance Mechanism Toolkit
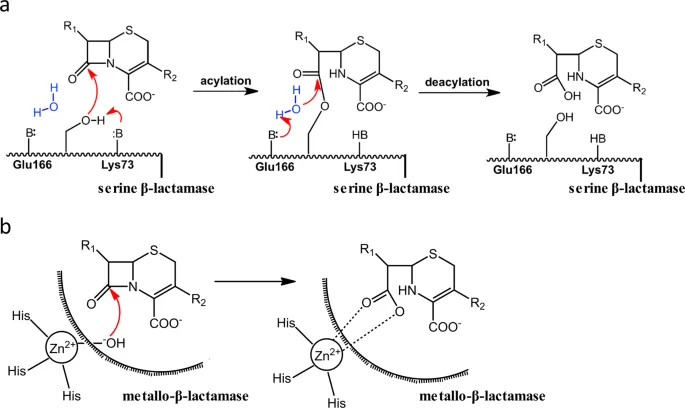
Enzymatic Inactivation: The Chemical Warfare Division
Enzymatic resistance represents the most clinically significant mechanism for antimicrobial inactivation:
-
β-Lactamase Classification (Ambler)
- Class A (serine): TEM, SHV, CTX-M families
- Class B (metallo): NDM, VIM, IMP carbapenemases
- Class C (AmpC): chromosomal and plasmid-mediated
- Class D (OXA): oxacillinases and carbapenemases
- TEM variants: >200 described alleles
- CTX-M enzymes: >170 variants identified
- NDM carbapenemases: >30 variants reported
-
Kinetic Parameters
- TEM-1 vs ampicillin: kcat = 2,000 s⁻¹, Km = 20 μM
- CTX-M-15 vs ceftriaxone: kcat = 150 s⁻¹, Km = 15 μM
- NDM-1 vs meropenem: kcat = 25 s⁻¹, Km = 30 μM
- Catalytic efficiency: 10⁵ to 10⁷ M⁻¹s⁻¹
- Substrate spectrum: 5-50 different antibiotics
- Expression levels: 10³-10⁶ molecules per cell
📌 Remember: BLADE - Beta-Lactamases Always Destroy Everything (broad spectrum activity)
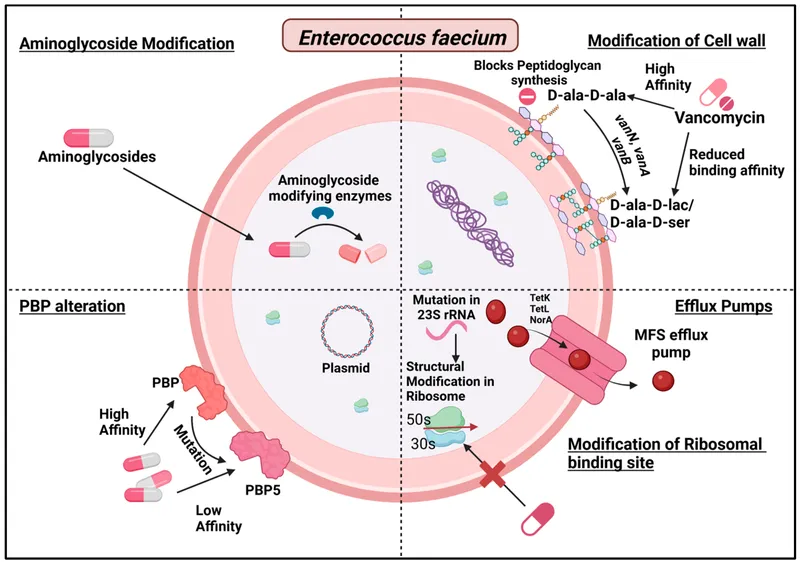
Target Modification: The Molecular Camouflage System
Target modification mechanisms alter antimicrobial binding sites while preserving essential cellular functions:
| Resistance Type | Target Modification | Frequency | Clinical Impact |
|---|---|---|---|
| PBP alterations | Low-affinity variants | 60-90% MRSA | β-lactam resistance |
| 16S rRNA methylation | A1408 methylation | 15-30% isolates | Aminoglycoside resistance |
| DNA gyrase mutations | Ser83/Asp87 changes | 40-80% resistance | Quinolone resistance |
| 23S rRNA mutations | A2058G substitution | 20-50% isolates | Macrolide resistance |
| Folate pathway | DHFR/DHPS variants | 70-95% resistance | Trimethoprim/sulfonamide |
- Point mutations: single nucleotide changes
- Domain swapping: horizontal gene transfer
- Allelic replacement: homologous recombination
- MRSA mecA: PBP2a with 1000-fold reduced affinity
- VRE vanA: D-Ala-D-Lac instead of D-Ala-D-Ala
- Quinolone resistance: ParC/GyrA double mutations
⭐ Clinical Pearl: Target modification resistance typically requires 2-4 mutations for high-level resistance, explaining the stepwise increase in MICs observed during treatment failure.
Efflux Pump Systems: The Cellular Bouncers
Efflux pumps actively remove antimicrobials from bacterial cells before they reach therapeutic concentrations:
-
Pump Classification
- ABC transporters: ATP-dependent, broad spectrum
- MFS pumps: proton-motive force, specific substrates
- RND pumps: tripartite systems, gram-negative
- AcrAB-TolC (E. coli): >100 different substrates
- MexAB-OprM (P. aeruginosa): β-lactams, quinolones
- NorA (S. aureus): quinolones, ethidium bromide
-
Clinical Resistance Levels
- Single pump overexpression: 4-16 fold MIC increase
- Multiple pump activation: 32-128 fold MIC increase
- Regulatory mutations: constitutive expression
- marA overexpression: 5-10 fold resistance increase
- mexR mutations: 20-50 fold pump upregulation
- acrR deletions: 10-25 fold efflux enhancement
💡 Master This: Bacterial resistance mechanisms operate as integrated defense networks where multiple mechanisms combine to create synergistic resistance levels that exceed the sum of individual contributions.
Understanding resistance mechanisms reveals how bacteria defeat antimicrobials at the molecular level. Clinical resistance patterns demonstrate how these mechanisms manifest in patient care settings.
🛡️ Molecular Defense Arsenal: The Resistance Mechanism Toolkit
🏥 Clinical Resistance Battlegrounds: The Hospital Ecosystem

Resistance Surveillance Architecture
Clinical resistance monitoring requires systematic data collection and analysis across multiple healthcare dimensions:
-
Surveillance Metrics
- Resistance prevalence: % resistant isolates per pathogen
- Resistance density: resistant isolates per 1000 patient-days
- Resistance trends: annual % change in resistance rates
- MRSA prevalence: 15-60% across institutions
- ESBL-producing E. coli: 10-40% in most regions
- Carbapenem-resistant Enterobacteriaceae: 1-15% emerging
-
Risk Stratification Factors
- ICU vs ward: 2-5 fold higher resistance rates
- Length of stay: +5% resistance risk per day
- Prior antibiotic exposure: 10-50% increased risk
- Fluoroquinolone exposure: 3-8 fold MRSA risk
- Carbapenem exposure: 5-15 fold CRE risk
- Broad-spectrum β-lactams: 2-6 fold ESBL risk
📌 Remember: TRACK - Trends Reveal Antibiotic Choice Keys (surveillance guides therapy)
High-Priority Resistance Patterns
Clinical resistance patterns follow predictable epidemiological distributions that guide empirical therapy decisions:
| Pathogen | Resistance Mechanism | Prevalence Range | Treatment Impact |
|---|---|---|---|
| MRSA | mecA-mediated | 15-60% | β-lactam failure |
| VRE | vanA/vanB genes | 5-30% | Glycopeptide failure |
| ESBL-EC | CTX-M enzymes | 10-40% | Cephalosporin failure |
| CRE | Carbapenemases | 1-15% | Carbapenem failure |
| MDR-PA | Multiple mechanisms | 20-50% | Limited options |
- Northern Europe: <5% ESBL prevalence
- Southern Europe: 15-25% ESBL prevalence
- Asia-Pacific: 30-60% ESBL prevalence
- CTX-M-15: dominant in Europe/North America
- CTX-M-14: prevalent in Asia
- NDM carbapenemases: endemic in South Asia
⭐ Clinical Pearl: Resistance rates >20% for any pathogen-antibiotic combination typically exclude that antibiotic from empirical therapy guidelines, following the "20% rule" in antimicrobial stewardship.
Resistance Transmission Dynamics
Healthcare-associated resistance transmission follows mathematical models that predict outbreak patterns:
-
Basic Reproduction Number (R₀)
- MRSA transmission: R₀ = 1.2-2.5 in ICU settings
- VRE transmission: R₀ = 1.5-3.0 in high-risk units
- CRE transmission: R₀ = 2.0-4.0 without intervention
- Hand hygiene compliance: reduces R₀ by 40-60%
- Contact precautions: reduces R₀ by 50-80%
- Environmental cleaning: reduces R₀ by 20-40%
-
Transmission Pathways
- Direct patient contact: 60-80% of transmissions
- Environmental contamination: 15-25% of transmissions
- Healthcare worker hands: 10-20% of transmissions
- Environmental survival: days to months
- Contaminated surfaces: 20-60% in patient rooms
- Medical equipment: 10-40% contamination rates
💡 Master This: Clinical resistance patterns emerge from predictable epidemiological forces where antimicrobial selection pressure, transmission dynamics, and infection control practices create mathematical relationships that enable resistance forecasting and intervention targeting.
Clinical resistance patterns provide the foundation for understanding how genetic mechanisms translate into patient care challenges. Resistance evolution reveals the long-term trajectory of antimicrobial effectiveness.
🏥 Clinical Resistance Battlegrounds: The Hospital Ecosystem
🔬 Resistance Evolution Engine: The Adaptive Machinery
Evolutionary Pressure Dynamics
Antimicrobial selection pressure creates predictable evolutionary trajectories that follow mathematical principles:
-
Selection Coefficient Calculations
- Fitness advantage: s = (Wresistant - Wsusceptible) / Wsusceptible
- Typical values: s = 0.1-0.8 under antibiotic pressure
- Population dynamics: exponential growth of resistant variants
- Doubling time reduction: 20-50% for resistant strains
- Competitive advantage: 2-10 fold under selection
- Fixation time: 10-50 generations depending on s-value
-
Mutation Rate Contributions
- Spontaneous mutations: 10⁻⁸ to 10⁻¹⁰ per base pair per generation
- Hypermutator strains: 10-1000 fold increased mutation rates
- Stress-induced mutagenesis: SOS response activation
- DNA polymerase errors: 10⁻⁴ to 10⁻⁶ per replication
- Recombination events: 10⁻³ to 10⁻⁵ per generation
- Horizontal transfer: 10⁻⁵ to 10⁻⁷ per cell per generation
📌 Remember: EVOLVE - Every Variant Outcompetes Less Viable Enemies (selection drives resistance)
Resistance Emergence Patterns
Resistance evolution follows predictable temporal and spatial patterns that enable forecasting:
| Resistance Type | Emergence Time | Spread Pattern | Persistence |
|---|---|---|---|
| Point mutations | Days to weeks | Clonal expansion | Stable |
| Plasmid acquisition | Hours to days | Horizontal spread | Variable |
| Transposon insertion | Days to weeks | Mixed patterns | Stable |
| Gene amplification | Hours to days | Unstable inheritance | Reversible |
| Regulatory changes | Days to weeks | Clonal expansion | Stable |
- First-step mutations: 2-8 fold MIC increase
- Second-step mutations: 8-32 fold MIC increase
- Third-step mutations: 32-128 fold MIC increase
- Quinolone resistance: gyrA → parC → efflux progression
- β-lactam resistance: PBP → β-lactamase → efflux combination
- Aminoglycoside resistance: target → enzyme → transport mechanisms
⭐ Clinical Pearl: Resistance evolution typically requires 2-4 independent mutations for high-level resistance, explaining why combination therapy reduces resistance emergence by 100-1000 fold.
Population Genetics of Resistance
Bacterial populations exhibit complex genetic structures that influence resistance evolution and spread:
-
Clonal Complex Analysis
- Sequence types (ST): multilocus sequence typing
- Clonal complexes: related ST groups
- Pandemic clones: globally distributed lineages
- ST131 E. coli: CTX-M-15 ESBL producer
- ST258 K. pneumoniae: KPC carbapenemase carrier
- CC398 S. aureus: livestock-associated MRSA
-
Genetic Diversity Metrics
- Nucleotide diversity (π): 0.001-0.01 for most species
- Tajima's D: selection vs neutrality indicator
- Recombination rates: 10⁻⁶ to 10⁻⁴ per site per generation
- High recombination: S. pneumoniae, N. gonorrhoeae
- Moderate recombination: E. coli, S. aureus
- Low recombination: M. tuberculosis, S. typhi
💡 Master This: Resistance evolution operates as a continuous adaptive process where antimicrobial pressure, genetic variation, and population dynamics create predictable evolutionary trajectories that can be modeled mathematically and targeted through strategic interventions.
Understanding resistance evolution provides the foundation for predicting future resistance trends and designing sustainable antimicrobial strategies that account for bacterial adaptive capacity.
🔬 Resistance Evolution Engine: The Adaptive Machinery
Practice Questions: Microbial genetics and drug resistance
Test your understanding with these related questions
A 42-year-old woman with a history of multiple sclerosis and recurrent urinary tract infections comes to the emergency department because of flank pain and fever. Her temperature is 38.8°C (101.8°F). Examination shows left-sided costovertebral angle tenderness. She is admitted to the hospital and started on intravenous vancomycin. Three days later, her symptoms have not improved. Urine culture shows growth of Enterococcus faecalis. Which of the following best describes the most likely mechanism of antibiotic resistance in this patient?










