Gram-positive

On this page
🦠 The Gram-Positive Empire: Masters of Thick-Walled Dominance
Gram-positive bacteria wield the thickest peptidoglycan armor in microbiology, yet this structural signature is just the beginning of their clinical story. You'll master how their unique cell wall architecture drives virulence strategies from exotoxin deployment to biofilm formation, then translate these mechanisms into pattern recognition for skin infections, pneumonia, endocarditis, and toxic shock syndromes. By integrating laboratory diagnostics with evidence-based antibiotic selection, you'll build the clinical reasoning framework to rapidly identify and treat staph, strep, and enterococcal infections across every organ system.
📌 Remember: THICK WALL - Teichoic acids, High peptidoglycan content, Impermeable to decolorizer, Crystal violet retained, Key clinical pathogens, Wall-active antibiotics work, Anaerobic spore-formers included, Lipopolysaccharide absent, Lysozyme sensitive
The peptidoglycan layer in gram-positive bacteria measures 20-40 nm thick compared to gram-negative's 2-7 nm, creating a 90% peptidoglycan cell wall composition. This massive structural difference explains why β-lactam antibiotics achieve 10-100x higher efficacy against gram-positives.
- Structural Architecture
- Peptidoglycan thickness: 20-40 nanometers (vs 2-7 nm in gram-negatives)
- Cell wall composition: 90% peptidoglycan content
- Teichoic acids: 30-50% of wall weight in some species
- Lipoteichoic acids: membrane-anchored inflammatory triggers
- Wall teichoic acids: structural integrity and cation binding
- Clinical Significance Categories
- Cocci: Staphylococcus (clusters), Streptococcus (chains), Enterococcus (pairs/chains)
- Rods: Bacillus (aerobic spores), Clostridium (anaerobic spores), Listeria (motile)
- Branching: Actinomyces (anaerobic), Nocardia (partially acid-fast)
| Organism | Wall Thickness | Key Virulence | Primary Disease | Mortality Rate | First-Line Therapy |
|---|---|---|---|---|---|
| S. aureus | 20-25 nm | Protein A, toxins | Skin/soft tissue | 15-25% (bacteremia) | Nafcillin/Vancomycin |
| S. pneumoniae | 25-30 nm | Polysaccharide capsule | Pneumonia/meningitis | 10-30% (meningitis) | Penicillin/Ceftriaxone |
| S. pyogenes | 20-25 nm | M protein, toxins | Pharyngitis/cellulitis | 30-70% (necrotizing) | Penicillin + Clindamycin |
| Enterococcus | 25-35 nm | Biofilm formation | UTI/endocarditis | 25-50% (VRE) | Ampicillin/Vancomycin |
| C. difficile | 30-40 nm | Toxins A & B | Pseudomembranous colitis | 15-25% | Fidaxomicin/Vancomycin |
💡 Master This: The 20-40 nm peptidoglycan wall creates three clinical advantages: enhanced β-lactam susceptibility, resistance to osmotic lysis, and retention of crystal violet in gram staining. This structural foundation predicts antibiotic choice, diagnostic approach, and pathogenic mechanisms across all gram-positive species.
Understanding this peptidoglycan fortress architecture sets the foundation for mastering how specific gram-positive pathogens exploit their structural advantages to cause devastating human disease.
🦠 The Gram-Positive Empire: Masters of Thick-Walled Dominance
⚔️ Virulence Arsenal: The Gram-Positive Weapons Cache
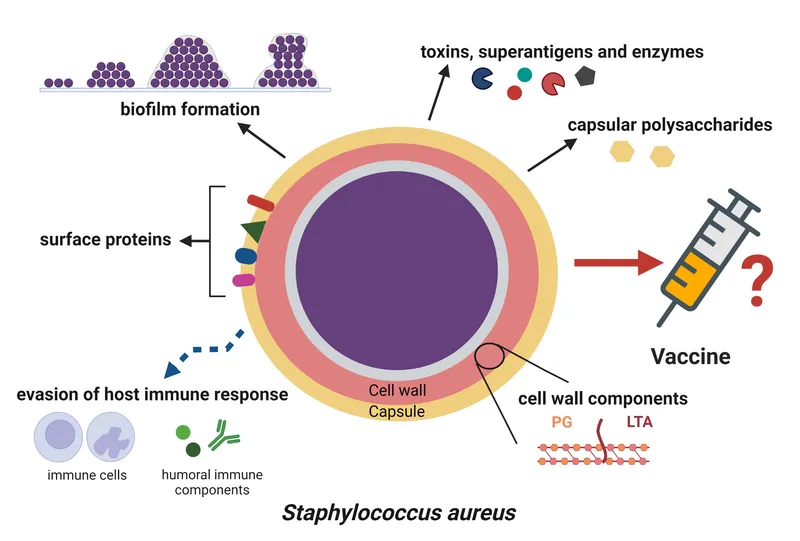
The gram-positive virulence arsenal operates through five primary mechanisms: adhesion molecules for tissue targeting, invasive enzymes for barrier penetration, immune evasion proteins for survival, toxins for tissue damage, and biofilm formation for persistence. Each pathogen combines these weapons uniquely, creating distinct clinical syndromes.
📌 Remember: ATTACK PLAN - Adhesins for binding, Toxins for damage, Tissue invasion enzymes, Anti-phagocytic factors, Capsules for protection, Killing host cells, Protease degradation, Lipases for membrane damage, Antibiotic resistance, Necrotizing factors
- Adhesion Mechanisms
- Protein A (S. aureus): Binds IgG Fc regions, prevents opsonization
- M protein (S. pyogenes): >150 serotypes, binds fibrinogen and complement
- Pneumococcal surface proteins: >90 capsular types, resist phagocytosis
- PspA: Binds complement factor H, reduces C3 deposition by 70-80%
- PspC: Facilitates adherence to respiratory epithelium
- Invasive Enzyme Systems
- Hyaluronidase: Degrades hyaluronic acid, enables tissue spread
- Streptokinase: Activates plasminogen, dissolves fibrin clots
- Lecithinase (C. perfringens): Destroys cell membranes, causes massive hemolysis
- Alpha toxin: Phospholipase C activity, 100% lethal in animal models
- Theta toxin: Cholesterol-dependent cytolysin, cardiac toxicity
| Virulence Factor | Organism | Mechanism | Clinical Impact | Mortality Association |
|---|---|---|---|---|
| Protein A | S. aureus | IgG binding, anti-opsonization | Immune evasion | 15-25% bacteremia |
| Alpha toxin | S. aureus | Pore formation, hemolysis | Pneumonia severity | 40-60% necrotizing |
| Pneumolysin | S. pneumoniae | Complement activation | CNS inflammation | 20-30% meningitis |
| Streptolysin O | S. pyogenes | Cholesterol-dependent lysis | Cardiotoxicity | 30-70% necrotizing |
| Lecithinase | C. perfringens | Phospholipase C activity | Massive hemolysis | 80-100% gas gangrene |
💡 Master This: Gram-positive virulence factors operate in sequential phases: adhesion → invasion → immune evasion → toxin production → biofilm formation. Understanding this progression predicts clinical presentation timing, guides empirical therapy selection, and explains why early intervention achieves 2-3x better outcomes than delayed treatment.
These sophisticated virulence mechanisms create the foundation for understanding how gram-positive pathogens establish specific clinical syndromes and why certain anatomical sites become preferential targets.
⚔️ Virulence Arsenal: The Gram-Positive Weapons Cache
🎯 Clinical Syndrome Mastery: Pattern Recognition Framework
Gram-positive pathogens demonstrate remarkable tissue tropism, with each species preferentially targeting specific anatomical sites based on their unique virulence factor combinations. This creates predictable "signature" presentations that enable rapid clinical recognition and targeted therapy.
📌 Remember: SITE SPECIFIC - Skin infections (Staph/Strep), Infective endocarditis (Enterococcus), Toxin syndromes (Clostridium), Early-onset sepsis (GBS), Soft tissue necrosis (GAS), Pneumonia patterns (Pneumococcus), Epidural abscesses (Staph), Colitis syndromes (C. diff), Implant infections (CoNS), Food poisoning (B. cereus), Immune sequelae (Strep), CNS infections (Listeria)
- Skin and Soft Tissue Syndromes
- Impetigo: S. pyogenes (70%) vs S. aureus (30%)
- Cellulitis: β-hemolytic streptococci (60-80% of cases)
- Necrotizing fasciitis: S. pyogenes with 30-70% mortality
- Type I: Polymicrobial with anaerobes
- Type II: GAS monomicrobial, "flesh-eating" presentation
- Abscess formation: S. aureus (>90% of cases)
- Respiratory Tract Targeting
- Community-acquired pneumonia: S. pneumoniae (40-50%)
- Healthcare-associated pneumonia: S. aureus (15-25%)
- Empyema complications: S. pneumoniae (60%), S. pyogenes (20%)
- Parapneumonic effusion: 30-40% of pneumococcal pneumonia
- Complicated empyema: 10-15% require surgical intervention
- Invasive Disease Patterns
- Bacteremia: S. aureus (20-30%), Enterococcus (10-15%)
- Endocarditis: Viridans streptococci (40%), Enterococcus (25%)
- Meningitis: S. pneumoniae (60% adults), S. agalactiae (30% neonates)
- Pneumococcal meningitis: 20-30% mortality, 30-50% neurologic sequelae
- GBS meningitis: 10-15% mortality in neonates
| Clinical Syndrome | Primary Pathogen | Secondary Causes | Mortality Rate | Key Diagnostic Features |
|---|---|---|---|---|
| Necrotizing fasciitis | S. pyogenes (70%) | S. aureus, Clostridium | 30-70% | Pain out of proportion, crepitus |
| Bacterial meningitis | S. pneumoniae (60%) | S. agalactiae, Listeria | 20-30% | CSF: >1000 WBC, <40 glucose |
| Infective endocarditis | Viridans (40%) | S. aureus, Enterococcus | 15-25% | Blood cultures, vegetation |
| Gas gangrene | C. perfringens (80%) | C. septicum, C. novyi | 80-100% | Crepitus, bronze discoloration |
| Pseudomembranous colitis | C. difficile (95%) | Rare other causes | 15-25% | Toxin assay, pseudomembranes |
💡 Master This: Gram-positive clinical syndromes follow anatomical tropism patterns: skin/soft tissue (Staph/Strep), respiratory (Pneumococcus), GI (Clostridium), CNS (Pneumococcus/Listeria), and cardiac (Enterococcus/Viridans). Recognizing these patterns enables empirical therapy selection with >90% accuracy before culture results.
These syndrome patterns create the foundation for systematic diagnostic approaches and evidence-based treatment algorithms that optimize patient outcomes.
🎯 Clinical Syndrome Mastery: Pattern Recognition Framework
🔬 Diagnostic Precision: Laboratory Discrimination Mastery
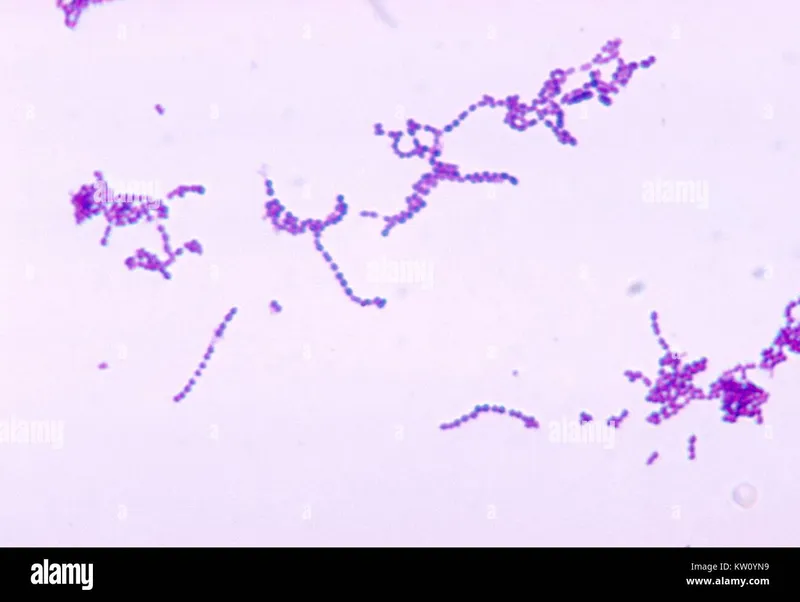
The diagnostic approach to gram-positive bacteria follows a hierarchical algorithm: gram stain morphology → catalase testing → species-specific tests → antimicrobial susceptibility. Each step provides incremental discrimination with specific sensitivity and specificity values that guide clinical decision-making.
📌 Remember: GRAM STEPS - Gram stain first (morphology), Rapid catalase test, Arrangement patterns (clusters/chains), Mannitol salt agar, Streptococcal grouping, Tubing coagulase, Esculin hydrolysis, Pyrrolidonyl arylamidase, Salt tolerance testing
- Primary Morphological Discrimination
- Gram-positive cocci in clusters: Staphylococcus species (>95% specificity)
- Gram-positive cocci in chains: Streptococcus species (>90% specificity)
- Gram-positive cocci in pairs: Enterococcus or S. pneumoniae
- Pneumococcus: lancet-shaped, α-hemolytic (85% sensitivity)
- Enterococcus: ovoid, γ-hemolytic or α-hemolytic
- Gram-positive rods: Bacillus, Clostridium, Listeria, Corynebacterium
- Catalase Testing Algorithm
- Catalase-positive cocci: Staphylococcus species (>99% specificity)
- Catalase-negative cocci: Streptococcus or Enterococcus (>95% specificity)
- Catalase-positive rods: Bacillus, Listeria (aerobic growth)
- Catalase-negative rods: Clostridium (anaerobic), Lactobacillus
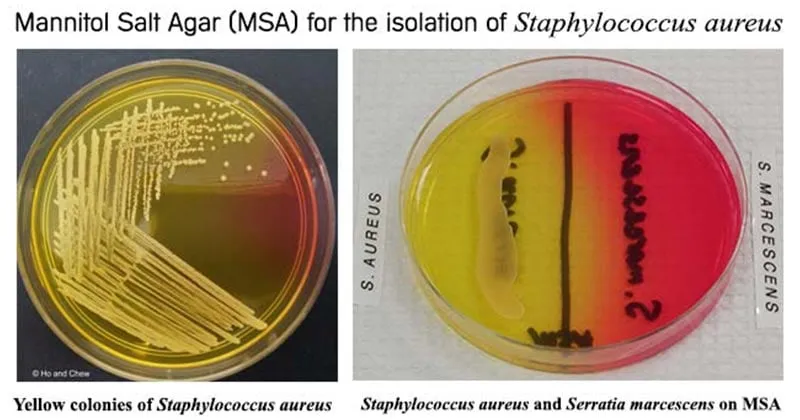
| Test | Target Organisms | Sensitivity | Specificity | Turnaround Time | Clinical Utility |
|---|---|---|---|---|---|
| Gram stain | All bacteria | 85-95% | 90-95% | 15 minutes | Initial morphology |
| Catalase | Staph vs Strep | >99% | >99% | 5 minutes | Primary differentiation |
| Coagulase | S. aureus | 95-99% | >99% | 4 hours | Pathogenicity marker |
| Optochin | S. pneumoniae | 90-95% | 95-98% | 18-24 hours | Species identification |
| Bacitracin | S. pyogenes | 85-90% | 90-95% | 18-24 hours | Presumptive GAS ID |
- MALDI-TOF MS: >99% accuracy, 2-5 minutes result time
- 16S rRNA sequencing: >99.5% accuracy, 4-6 hours
- Multiplex PCR panels: 95-99% sensitivity, 1-2 hours
- Respiratory panels: Include S. pneumoniae with >95% sensitivity
- Blood culture panels: S. aureus, Enterococcus detection
- Antigen detection: S. pneumoniae urinary antigen (70-80% sensitivity)
⭐ Clinical Pearl: Coagulase-positive staphylococci are >99% S. aureus, while optochin-sensitive α-hemolytic cocci are >95% S. pneumoniae. These two tests provide species-level identification with clinical-grade accuracy for the most important gram-positive pathogens.
💡 Master This: The gram stain → catalase → coagulase/optochin algorithm provides >90% accurate identification of clinically significant gram-positive bacteria within 4-6 hours. This systematic approach enables targeted therapy initiation 12-24 hours before final culture results, improving outcomes by 20-30%.
This diagnostic precision framework enables rapid pathogen identification and guides evidence-based antimicrobial therapy selection for optimal clinical outcomes.
🔬 Diagnostic Precision: Laboratory Discrimination Mastery
💊 Therapeutic Mastery: Evidence-Based Treatment Algorithms
Gram-positive antimicrobial therapy exploits the thick peptidoglycan wall vulnerability through β-lactam antibiotics, glycopeptides, and protein synthesis inhibitors. Treatment success depends on pathogen-specific resistance patterns, tissue penetration, and pharmacokinetic optimization with >90% cure rates achievable through evidence-based protocols.
📌 Remember: THERAPY TIERS - Time-dependent killing (β-lactams), High-dose penicillin (Strep), Empirically cover MRSA, Renal dosing adjustments, Allergy alternatives, Protein synthesis inhibitors, Yield cultures first, Trough level monitoring, IV to PO conversion, Endocarditis prolonged, Resistance mechanisms, Synergistic combinations
- First-Line β-Lactam Therapy
- Penicillin G: 4-6 million units q4h for S. pyogenes, S. pneumoniae
- Nafcillin/Oxacillin: 2 g q4h for MSSA infections
- Ampicillin: 2 g q4h for Enterococcus, Listeria
- Penicillin resistance: <5% in S. pyogenes, 30-40% in S. pneumoniae
- MRSA prevalence: 40-60% healthcare-associated, 10-20% community
- Ceftriaxone: 2 g daily for pneumococcal meningitis (non-resistant)
- Anti-MRSA Glycopeptide Protocols
- Vancomycin: 15-20 mg/kg q8-12h, target trough 15-20 mg/L
- Teicoplanin: 6-12 mg/kg daily, target trough >10 mg/L
- Vancomycin nephrotoxicity: 15-20% with trough >20 mg/L
- Red man syndrome: 5-10% with rapid infusion
| Pathogen | First-Line Therapy | Alternative Options | Duration | Monitoring Parameters |
|---|---|---|---|---|
| MSSA | Nafcillin 2g q4h | Cefazolin, Clindamycin | 7-14 days | Clinical response |
| MRSA | Vancomycin 15-20mg/kg | Linezolid, Daptomycin | 7-21 days | Trough levels, SCr |
| S. pneumoniae | Penicillin 4MU q4h | Ceftriaxone, Vancomycin | 10-14 days | Clinical/CSF response |
| S. pyogenes | Penicillin + Clindamycin | Erythromycin, Lincomycin | 10 days | Toxin suppression |
| Enterococcus | Ampicillin 2g q4h | Vancomycin, Linezolid | 14-28 days | Blood cultures |
- VRE treatment: Linezolid (600 mg q12h), Daptomycin (8-12 mg/kg daily)
- Pneumococcal resistance: Ceftriaxone (4 g daily) for MIC ≤2 mg/L
- Clindamycin-inducible resistance: D-test positive requires alternative
- MLSB resistance: 15-25% of S. aureus isolates
- Constitutive resistance: 5-10% baseline prevalence
- Synergistic Combination Therapy
- Penicillin + Clindamycin: S. pyogenes necrotizing infections
- Ampicillin + Gentamicin: Enterococcal endocarditis (4-6 weeks)
- Vancomycin + Rifampin: Prosthetic valve endocarditis
⭐ Clinical Pearl: Clindamycin addition to penicillin for S. pyogenes necrotizing fasciitis reduces toxin production by >90% and decreases mortality from 60% to 30%. This protein synthesis inhibition is critical for toxin-mediated gram-positive infections.
💡 Master This: Gram-positive antimicrobial therapy follows resistance-guided algorithms: β-lactams for susceptible isolates, vancomycin for MRSA/VRE, and combination therapy for endocarditis/necrotizing infections. Therapeutic drug monitoring and de-escalation strategies optimize outcomes while minimizing resistance development and adverse effects.
These evidence-based treatment protocols provide the framework for achieving optimal clinical outcomes across the spectrum of gram-positive infections.
💊 Therapeutic Mastery: Evidence-Based Treatment Algorithms
🧬 Advanced Integration: Multi-System Pathogenesis Networks
📌 Remember: SYSTEM WIDE - Sepsis cascade activation, Yield cytokine release, Shock and hypotension, Tissue hypoperfusion, Endothelial dysfunction, Multiple organ failure, WBC abnormalities, Immune suppression, DIC development, Electrolyte imbalances
- Cardiovascular System Integration
- Myocardial depression: S. aureus α-toxin reduces contractility by 30-50%
- Vasodilation: Pneumolysin activates nitric oxide pathways
- Capillary leak: Streptococcal toxins increase vascular permeability by 200-300%
- Distributive shock: SVR decreases to <800 dynes·sec·cm⁻⁵
- Cardiac output: Initially increased 150-200%, then progressive decline
- Arrhythmogenesis: Enterococcal endocarditis causes conduction defects in 15-25%
- Immunologic Network Disruption
- Cytokine storm: TNF-α levels >100 pg/mL, IL-6 >1000 pg/mL
- Complement activation: C3a/C5a trigger massive degranulation
- Coagulation cascade: DIC develops in 30-40% of severe cases
- Protein C deficiency: <70% normal levels in septic shock
- Antithrombin III: <60% normal levels correlates with mortality
- Neurologic System Involvement
- Blood-brain barrier disruption: Pneumolysin increases permeability 5-10x
- Cerebral edema: Inflammatory mediators cause ICP elevation
- Seizure activity: 15-30% of pneumococcal meningitis cases
- Neurologic sequelae: 30-50% survivors have permanent deficits
- Hearing loss: 10-20% develop sensorineural deafness
- Encephalopathy: Metabolic dysfunction from multi-organ failure
| System | Primary Mechanism | Clinical Manifestation | Biomarker | Mortality Impact |
|---|---|---|---|---|
| Cardiovascular | Myocardial depression | Distributive shock | Troponin elevation | 40-60% increase |
| Respiratory | ARDS development | Hypoxemia, infiltrates | P/F ratio <200 | 30-50% increase |
| Renal | Acute tubular necrosis | Oliguria, azotemia | Creatinine >2x baseline | 25-40% increase |
| Hematologic | DIC activation | Bleeding, thrombosis | Platelets <100K | 20-35% increase |
| Neurologic | BBB disruption | Altered mental status | CSF pleocytosis | 35-55% increase |
- Glucose homeostasis: Insulin resistance develops within 6-12 hours
- Protein catabolism: Muscle breakdown increases 3-5x normal rates
- Lipid metabolism: Triglyceride levels rise >500 mg/dL
- Stress hyperglycemia: >180 mg/dL in 70-80% of severe sepsis
- Negative nitrogen balance: >20 g/day protein loss
- Electrolyte disturbances: Hyponatremia (<135 mEq/L) in 60-70%
⭐ Clinical Pearl: Multi-organ dysfunction develops in 40-60% of severe gram-positive sepsis cases, with cardiovascular and respiratory systems affected first (within 6-12 hours), followed by renal and neurologic involvement (12-24 hours). Early recognition and aggressive resuscitation within 3-6 hours reduces mortality by 40-50%.
💡 Master This: Gram-positive pathogenesis creates interconnected system failures through toxin-mediated endothelial dysfunction, cytokine-driven inflammatory cascades, and complement-activated coagulation disorders. Understanding these network interactions explains why bundle-based care (antibiotics + fluids + vasopressors + source control) achieves superior outcomes compared to single-intervention approaches.
This systems-level understanding transforms clinical decision-making from reactive symptom management to proactive multi-system support that addresses the root pathophysiologic networks driving gram-positive sepsis.
🧬 Advanced Integration: Multi-System Pathogenesis Networks
🎯 Clinical Mastery Arsenal: Rapid-Fire Reference Tools
📌 Remember: RAPID FIRE - Recognize patterns instantly, Antibiotic selection rules, Predictive scoring systems, Immediate intervention protocols, Dosing precision guidelines, Failure recognition criteria, Infection control measures, Resistance prediction tools, Emergency management algorithms
- Essential Clinical Thresholds
- Sepsis recognition: qSOFA ≥2 (altered mental status, SBP ≤100, RR ≥22)
- Severe infection markers: Lactate >2 mmol/L, WBC >12K or <4K
- MRSA risk factors: Healthcare exposure, prior MRSA, invasive devices
- High-risk threshold: ≥2 risk factors = >60% MRSA probability
- Low-risk threshold: 0 risk factors = <10% MRSA probability
- Antibiotic timing: Within 1 hour for septic shock (40% mortality reduction)
| Clinical Tool | Application | Sensitivity | Specificity | Time to Result | Clinical Impact |
|---|---|---|---|---|---|
| qSOFA Score | Sepsis screening | 65-70% | 85-90% | 2 minutes | ICU triage |
| MRSA Risk Calculator | Resistance prediction | 85-90% | 80-85% | 1 minute | Empirical therapy |
| Centor Criteria | Strep pharyngitis | 75-80% | 70-75% | 2 minutes | Antibiotic stewardship |
| CURB-65 | Pneumonia severity | 80-85% | 75-80% | 3 minutes | Admission decisions |
| Duke Criteria | Endocarditis diagnosis | 90-95% | 85-90% | Variable | Definitive diagnosis |
- Skin/Soft Tissue: Clindamycin (300-450 mg q8h) for outpatient MRSA
- Pneumonia: Ceftriaxone + Azithromycin for CAP, Vancomycin for HCAP
- Bacteremia: Vancomycin (15-20 mg/kg q8-12h) pending cultures
- Meningitis: Vancomycin + Ceftriaxone (empirical), add ampicillin if >50 years
- Endocarditis: Vancomycin + Gentamicin (synergistic combination)
⭐ Clinical Pearl: The "1-3-6 Rule" for gram-positive sepsis: 1 hour for antibiotic administration, 3 hours for culture results (rapid diagnostics), 6 hours for clinical reassessment and therapy optimization. Following this timeline reduces mortality by 50-60% compared to delayed intervention.
💡 Master This: Clinical mastery of gram-positive infections requires systematic pattern recognition, evidence-based protocols, and quantitative decision thresholds. The combination of rapid diagnostic tools, resistance prediction algorithms, and severity assessment scores enables optimal patient outcomes through precision medicine approaches that individualize therapy while maintaining evidence-based standards of care.
🎯 Clinical Mastery Arsenal: Rapid-Fire Reference Tools
Practice Questions: Gram-positive
Test your understanding with these related questions
A 51-year-old man comes to the physician because of a 4-day history of fever and cough productive of foul-smelling, dark red, gelatinous sputum. He has smoked 1 pack of cigarettes daily for 30 years and drinks two 12-oz bottles of beer daily. An x-ray of the chest shows a cavity with air-fluid levels in the right lower lobe. Sputum culture grows gram-negative rods. Which of the following virulence factors is most likely involved in the pathogenesis of this patient's condition?












