Bacteria

On this page
🦠 The Bacterial Universe - Microscopic Masters of Survival
Bacteria are Earth's most successful organisms, thriving everywhere from deep-sea vents to human intestines, and understanding their survival strategies, metabolic versatility, and pathogenic mechanisms is essential for clinical medicine. You'll master pattern recognition frameworks that connect bacterial structure to disease presentation, learn how antimicrobial agents exploit specific vulnerabilities, and build integrated mental models linking microbiology to diagnosis and treatment across organ systems. This foundation transforms bacteria from abstract microbes into predictable adversaries you can identify, target, and defeat at the bedside.
The Bacterial Blueprint - Prokaryotic Perfection
Bacteria represent the ultimate minimalist design, packing essential life functions into cells 10-100 times smaller than eukaryotic counterparts. This streamlined architecture enables generation times as short as 20 minutes under optimal conditions, explaining their rapid adaptation and clinical significance.
- Size Range: 0.2-5.0 μm diameter
- Mycoplasma: 0.1-0.3 μm (smallest free-living organisms)
- E. coli: 1.0-1.5 μm × 2.0-6.0 μm
- Bacillus anthracis: 1.0-1.2 μm × 3.0-5.0 μm
- Genetic Material: Single circular chromosome (1-10 million base pairs)
- No nuclear membrane or histones
- Plasmids carry additional 1,000-200,000 base pairs
- 16S rRNA gene serves as universal bacterial identifier
📌 Remember: PRIM for prokaryotic features - Plasmids present, Ribosomes 70S, Introns absent, Membrane-bound organelles missing
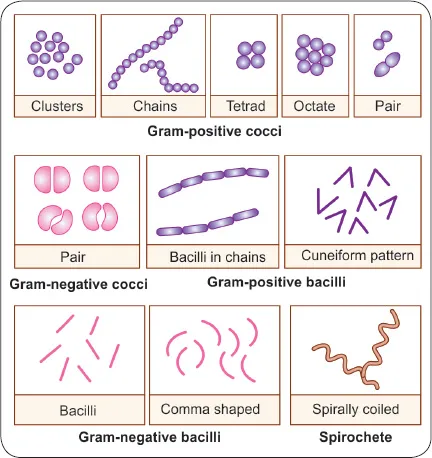
Morphological Mastery - Shape Determines Function
Bacterial morphology directly correlates with pathogenic mechanisms and clinical presentations. Understanding these relationships predicts virulence patterns and diagnostic approaches.
| Morphology | Examples | Size (μm) | Key Virulence | Clinical Significance |
|---|---|---|---|---|
| Cocci | Streptococcus | 0.5-1.0 | Chain formation | Pharyngitis, endocarditis |
| Bacilli | E. coli | 1.0 × 2.0 | Flagellar motility | UTI, sepsis |
| Spirilla | Campylobacter | 0.5 × 2.0 | Corkscrew motility | Gastroenteritis |
| Spirochetes | Treponema | 0.1 × 10 | Tissue penetration | Syphilis, CNS invasion |
| Pleomorphic | Mycoplasma | 0.1-0.3 | Cell wall absence | Atypical pneumonia |
- Arrangement Patterns:
- Diplococci: Pairs (Streptococcus pneumoniae)
- Tetrads: Groups of 4 (Micrococcus)
- Sarcinae: Cubes of 8 (Sarcina ventriculi)
- Streptococci: Chains (β-hemolytic Streptococcus)
- Staphylococci: Grape-like clusters (Staphylococcus aureus)
⭐ Clinical Pearl: Lancet-shaped diplococci in CSF indicate pneumococcal meningitis with >95% specificity, enabling immediate empirical therapy decisions
Essential Bacterial Components - The Survival Toolkit
Every bacterial structure serves specific survival and pathogenic functions. Mastering these relationships explains antibiotic targets and resistance mechanisms.
- Cell Wall Architecture:
- Peptidoglycan thickness: Gram-positive 20-80 nm vs. Gram-negative 2-7 nm
- Cross-linking: β-lactam antibiotics target transpeptidase enzymes
- Porins: Allow molecules <600 Da to pass through outer membrane
- Membrane Systems:
- Cytoplasmic membrane: 7-8 nm thick phospholipid bilayer
- Outer membrane: Gram-negative exclusive, contains lipopolysaccharide
- Periplasmic space: 12-15 nm gap containing degradative enzymes
💡 Master This: Cell wall differences determine antibiotic susceptibility patterns - Gram-positive bacteria show higher vancomycin sensitivity due to accessible peptidoglycan targets, while Gram-negative resistance stems from outer membrane barriers
Understanding bacterial fundamentals through this structural lens reveals the engineering principles behind microbial success, setting the foundation for comprehending complex pathogenic mechanisms and therapeutic strategies.
🦠 The Bacterial Universe — Microscopic Masters of Survival
⚡ Metabolic Powerhouses - Bacterial Energy Systems
Energy Generation Strategies - Metabolic Flexibility
Bacteria employ diverse energy-harvesting mechanisms, explaining their ecological success and clinical significance. These pathways determine growth characteristics and diagnostic profiles.
- Respiratory Classifications:
- Obligate aerobes: Require >15% O₂ (Mycobacterium tuberculosis)
- Facultative anaerobes: Grow with/without O₂ (E. coli, Staphylococcus)
- Obligate anaerobes: Die at >0.5% O₂ (Clostridium, Bacteroides)
- Microaerophiles: Require 2-10% O₂ (Campylobacter jejuni)
- Aerotolerant: Indifferent to O₂ (Enterococcus faecalis)
📌 Remember: FOAM for oxygen requirements - Facultative (most pathogens), Obligate aerobes (TB), Anaerobes (wound infections), Microaerophiles (GI pathogens)
Biochemical Identification Arsenal - Metabolic Fingerprints
Bacterial identification relies on characteristic metabolic patterns that remain consistent across strains. These biochemical signatures enable rapid species-level identification.
| Test | Principle | Positive Examples | Clinical Significance |
|---|---|---|---|
| Catalase | H₂O₂ → H₂O + O₂ | Staphylococcus, Bacillus | Differentiates from Streptococcus |
| Oxidase | Cytochrome c oxidase | Pseudomonas, Neisseria | Gram-negative identification |
| Coagulase | Fibrinogen → Fibrin | S. aureus | Pathogenic staphylococci |
| Indole | Tryptophan → Indole | E. coli, Proteus | Enterobacteriaceae speciation |
| Urease | Urea → NH₃ + CO₂ | Proteus, H. pylori | Rapid identification |
- Fermentation Patterns:
- Glucose fermentation: Universal among Enterobacteriaceae
- Lactose fermentation: E. coli (positive) vs. Salmonella (negative)
- Mannitol fermentation: S. aureus (positive) vs. S. epidermidis (negative)
- Sucrose fermentation: Vibrio cholerae (positive, 6-hour rapid test)
⭐ Clinical Pearl: Positive urease test within 4 hours indicates Proteus species with >98% accuracy, explaining the characteristic "fishy" odor and alkaline urine pH in UTIs
Specialized Metabolic Pathways - Unique Signatures
Certain bacteria possess distinctive metabolic capabilities that serve as diagnostic markers and explain specific clinical presentations.
- Unique Enzyme Systems:
- β-lactamase production: >50% of S. aureus strains
- Lecithinase (phospholipase C): Clostridium perfringens
- Hyaluronidase: Group A Streptococcus "spreading factor"
- Streptokinase: Fibrinolytic enzyme in S. pyogenes
- Specialized Growth Requirements:
- X factor (hemin) + V factor (NAD): Haemophilus influenzae
- Chocolate agar: Required for fastidious organisms
- 5-10% CO₂: Enhances growth of Streptococcus pneumoniae
- 35°C optimal: Most human pathogens vs. 25°C environmental bacteria
💡 Master This: Metabolic requirements directly correlate with pathogenic potential - fastidious organisms requiring complex nutrients typically show higher virulence and tissue specificity, explaining why H. influenzae causes meningitis while environmental bacteria rarely invade sterile sites
These metabolic principles form the foundation for understanding bacterial behavior in clinical specimens, guiding both diagnostic approaches and therapeutic targeting strategies.
⚡ Metabolic Powerhouses — Bacterial Energy Systems
🔬 Diagnostic Mastery - Pattern Recognition Frameworks
The Gram Stain Algorithm - Primary Classification Gateway
Gram staining provides the fundamental branching point for all bacterial identification, with >95% reproducibility when performed correctly. This single test narrows diagnostic possibilities from thousands to dozens.
- Gram-Positive Patterns (Purple/Blue):
- Cocci in clusters: Think Staphylococcus → Catalase test
- Catalase positive: S. aureus vs. coagulase-negative staphylococci
- Coagulase positive: S. aureus (>99% specificity)
- Cocci in chains: Think Streptococcus → Hemolysis pattern
- β-hemolytic: Groups A, B, C, G
- α-hemolytic: S. pneumoniae, viridans group
- γ-hemolytic: Enterococcus species
- Rods (bacilli): Spore-forming vs. non-spore-forming
- Spore-formers: Bacillus, Clostridium
- Non-spore-formers: Listeria, Corynebacterium
- Cocci in clusters: Think Staphylococcus → Catalase test
📌 Remember: SCAB for Gram-positive morphology - Staphylococcus (clusters), Corynebacterium (rods), Actinomyces (branching), Bacillus (spores)
Gram-Negative Identification Matrix - Systematic Approach
Gram-negative bacteria require multi-step identification algorithms based on morphology, oxidase reaction, and biochemical profiles.
| Morphology | Oxidase | Key Tests | Major Pathogens | Clinical Syndromes |
|---|---|---|---|---|
| Cocci | Positive | Sugar fermentation | Neisseria species | Meningitis, gonorrhea |
| Rods | Positive | Glucose fermentation | Pseudomonas | Healthcare-associated infections |
| Rods | Negative | Lactose fermentation | Enterobacteriaceae | UTI, gastroenteritis |
| Curved | Positive | Growth at 42°C | Campylobacter | Bloody diarrhea |
| Spiral | Variable | Dark-field microscopy | Spirochetes | Syphilis, Lyme disease |
| %%{init: {'flowchart': {'htmlLabels': true}}}%% | ||||
| flowchart TD |
Start["<b>🧫 Gram-Negative</b><br><span style='display:block; text-align:left; color:#555'>• Routine screening</span><span style='display:block; text-align:left; color:#555'>• Pathogen ID</span>"]
MacNo["<b>🔬 Limited Growth</b><br><span style='display:block; text-align:left; color:#555'>• Chocolate agar (+)</span><span style='display:block; text-align:left; color:#555'>• MacConkey (-)</span>"]
MacYes["<b>🔬 Good Growth</b><br><span style='display:block; text-align:left; color:#555'>• Multi-agar growth</span><span style='display:block; text-align:left; color:#555'>• Includes MacConkey</span>"]
Cocci["<b>🩺 Cocci Morphology</b><br><span style='display:block; text-align:left; color:#555'>• Coccobacilli forms</span><span style='display:block; text-align:left; color:#555'>• Small rounded cells</span>"]
Sicca["<b>✅ N. sicca</b><br><span style='display:block; text-align:left; color:#555'>• Neisseria species</span><span style='display:block; text-align:left; color:#555'>• No isolation agar</span>"]
Flu["<b>✅ H. influenzae</b><br><span style='display:block; text-align:left; color:#555'>• Isolation agar (+)</span><span style='display:block; text-align:left; color:#555'>• Growth on selective</span>"]
Rods["<b>🩺 Rod Morphology</b><br><span style='display:block; text-align:left; color:#555'>• Gram-negative rods</span><span style='display:block; text-align:left; color:#555'>• Bacilli shape</span>"]
LacPos["<b>📋 Lactose Ferment (+)</b><br><span style='display:block; text-align:left; color:#555'>• Fast fermenters</span><span style='display:block; text-align:left; color:#555'>• Color change observed</span>"]
LacNeg["<b>📋 Lactose Ferment (-)</b><br><span style='display:block; text-align:left; color:#555'>• Non-fermenters</span><span style='display:block; text-align:left; color:#555'>• No color change</span>"]
Ecoli["<b>✅ E. coli</b><br><span style='display:block; text-align:left; color:#555'>• Urease/Citrate (-)</span><span style='display:block; text-align:left; color:#555'>• Common coliform</span>"]
Kleb["<b>✅ K. pneumoniae</b><br><span style='display:block; text-align:left; color:#555'>• Urease/Citrate (+)</span><span style='display:block; text-align:left; color:#555'>• Mucoid colonies</span>"]
Entero["<b>✅ E. aerogenes</b><br><span style='display:block; text-align:left; color:#555'>• Urease (-)/Citrate (+)</span><span style='display:block; text-align:left; color:#555'>• Enteric pathogen</span>"]
Pseud["<b>✅ P. aeruginosa</b><br><span style='display:block; text-align:left; color:#555'>• Oxidase (+)/SIM (-)</span><span style='display:block; text-align:left; color:#555'>• Oppourtunistic</span>"]
Prot["<b>✅ P. vulgaris</b><br><span style='display:block; text-align:left; color:#555'>• Oxidase (-)/SIM (+)</span><span style='display:block; text-align:left; color:#555'>• Swarming motility</span>"]
Start --> MacNo
Start --> MacYes
MacNo --> Cocci
Cocci -->|No Iso Agar| Sicca
Cocci -->|Iso Agar +| Flu
MacYes --> Rods
Rods --> LacPos
Rods --> LacNeg
LacPos --> Ecoli
LacPos --> Kleb
LacPos --> Entero
LacNeg -->|Oxidase +| Pseud
LacNeg -->|Oxidase -| Prot
style Start fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8
style MacNo fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C
style MacYes fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C
style Cocci fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8
style Sicca fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Flu fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Rods fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8
style LacPos fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E
style LacNeg fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E
style Ecoli fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Kleb fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Entero fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Pseud fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
style Prot fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
* **Enterobacteriaceae Differentiation**:
- **Lactose fermenters**: E. coli, Klebsiella, Enterobacter
+ **Indole positive**: E. coli (**>90%** of strains)
+ **Indole negative**: Klebsiella pneumoniae
- **Lactose non-fermenters**: Salmonella, Shigella, Proteus
+ **H₂S production**: Salmonella (positive), Shigella (negative)
+ **Motility**: Salmonella (motile), Shigella (non-motile)
> ⭐ **Clinical Pearl**: **Oxidase-positive, glucose-non-fermenting** Gram-negative rods in respiratory specimens indicate **Pseudomonas aeruginosa** with **>95% probability**, requiring immediate anti-pseudomonal antibiotic coverage
### Rapid Identification Technologies - Modern Diagnostic Arsenal
Contemporary bacterial identification employs automated systems and molecular methods that provide results within **2-6 hours** compared to traditional **24-48 hour** methods.
* **Automated Systems**:
- **VITEK 2**: **4-8 hour** identification with **>95% accuracy**
- **Phoenix**: Combines identification with susceptibility testing
- **MicroScan**: Overnight incubation with morning results
* **Molecular Methods**:
- **16S rRNA sequencing**: Universal bacterial identifier
- **MALDI-TOF MS**: **<5 minute** identification from colonies
- **PCR-based systems**: **2-4 hour** results from positive blood cultures
- **Peptide nucleic acid probes**: Rapid species confirmation
> 💡 **Master This**: **MALDI-TOF mass spectrometry** identifies bacteria by analyzing **ribosomal protein fingerprints**, achieving **>97% accuracy** within **5 minutes** of colony analysis, revolutionizing clinical microbiology workflow and enabling same-day organism identification
These diagnostic frameworks transform bacterial identification from memorization-based guesswork into systematic pattern recognition, enabling rapid, accurate clinical decision-making that directly improves patient outcomes.
🔬 Diagnostic Mastery — Pattern Recognition Frameworks
🎯 Antimicrobial Strategy - Precision Targeting Systems
Target-Based Antibiotic Classification - Precision Strike Points
Antibiotics achieve selectivity by targeting bacterial-specific structures and processes absent in human cells. Understanding these targets explains both therapeutic efficacy and resistance patterns.
- Cell Wall Synthesis Inhibitors:
- β-lactams: Target transpeptidase enzymes (PBPs)
- Penicillins: Narrow spectrum, MIC ≤0.12 μg/mL for susceptible organisms
- Cephalosporins: 4 generations with expanding spectrum
- Carbapenems: Broadest spectrum, last-resort agents
- Vancomycin: Binds D-Ala-D-Ala peptide termini
- MIC ≤2 μg/mL: Susceptible
- MIC 4-8 μg/mL: Intermediate resistance
- MIC ≥16 μg/mL: Resistant (VRE)
- β-lactams: Target transpeptidase enzymes (PBPs)
📌 Remember: CAMP for bactericidal antibiotics - Cephalosporins, Aminoglycosides, Metronidazole, Penicillins (plus fluoroquinolones, vancomycin)
Resistance Mechanism Matrix - Bacterial Defense Systems
Bacterial resistance develops through predictable mechanisms that determine treatment failure patterns and guide therapeutic choices.
| Mechanism | Examples | Affected Antibiotics | Clinical Impact |
|---|---|---|---|
| β-lactamase | ESBL, AmpC, KPC | β-lactams | >50% treatment failures |
| Target modification | MRSA (PBP2a) | Methicillin, oxacillin | >90% cross-resistance |
| Efflux pumps | P. aeruginosa | Multiple classes | 2-8 fold MIC increases |
| Permeability loss | Porin mutations | Carbapenems | >16 fold MIC increases |
| Enzymatic inactivation | Aminoglycoside modifying enzymes | Gentamicin, tobramycin | Complete resistance |
- Extended-Spectrum β-lactamases (ESBLs):
- Prevalence: >30% of E. coli, >20% of Klebsiella
- Hydrolysis: Penicillins, cephalosporins, aztreonam
- Inhibition: Clavulanic acid, tazobactam
- Treatment: Carbapenems preferred (>95% susceptibility)
⭐ Clinical Pearl: ESBL-producing organisms show in vitro susceptibility to cephalosporins but clinical failure rates >40% due to inoculum effects and resistance induction, mandating carbapenem therapy for serious infections
Pharmacokinetic-Pharmacodynamic Optimization - Dosing Science
Antibiotic efficacy depends on achieving optimal drug concentrations at infection sites while minimizing toxicity and resistance selection.
- PK/PD Parameters:
- Time-dependent killing: β-lactams require T>MIC >40-70% of dosing interval
- Concentration-dependent killing: Aminoglycosides need Cmax/MIC >8-10
- AUC-dependent killing: Fluoroquinolones require AUC/MIC >125
- Tissue Penetration:
- CNS penetration: <10% for most antibiotics
- Bone penetration: Fluoroquinolones >60%, β-lactams <20%
- Intracellular penetration: Macrolides >90%, β-lactams <5%
- Renal Adjustment:
- CrCl <50 mL/min: Reduce aminoglycoside, vancomycin doses
- CrCl <30 mL/min: Extend β-lactam dosing intervals
- Dialysis: 50-70% drug removal for water-soluble antibiotics
💡 Master This: Combination therapy achieves synergy through complementary mechanisms - β-lactam + aminoglycoside combinations show 2-4 log greater bacterial killing than either agent alone, explaining improved outcomes in Enterococcus endocarditis and Pseudomonas bacteremia
These antimicrobial principles enable rational drug selection that maximizes therapeutic efficacy while minimizing resistance development, transforming empirical prescribing into precision medicine approaches.
🎯 Antimicrobial Strategy — Precision Targeting Systems
🌐 Pathogenic Networks - Multi-System Integration
Virulence Factor Integration - The Pathogenic Arsenal
Successful bacterial pathogens coordinate multiple virulence mechanisms that work synergistically to establish infection, evade host defenses, and cause tissue damage.
- Adhesion and Colonization:
- Pili and fimbriae: Type 1 fimbriae bind mannose residues on uroepithelial cells
- Adhesins: >50 different types in E. coli alone
- Biofilm formation: 1000-fold increased antibiotic resistance
- Quorum sensing: Coordinates virulence at 10⁶-10⁸ CFU/mL densities
- Immune Evasion Strategies:
- Capsule formation: >90 serotypes in S. pneumoniae
- Antigenic variation: >1000 variants in Salmonella flagellar antigens
- Complement resistance: Factor H binding by N. meningitidis
- Intracellular survival: Phagosome-lysosome fusion inhibition
📌 Remember: CITE for major virulence categories - Capsules (immune evasion), Invasins (tissue penetration), Toxins (tissue damage), Enzymes (tissue breakdown)
Host-Pathogen Interaction Networks - Dynamic Battlegrounds
Bacterial infections represent dynamic interactions between pathogen virulence factors and host immune responses, with outcomes determined by the balance of these competing forces.
| Host Factor | Bacterial Counter | Clinical Outcome | Therapeutic Target |
|---|---|---|---|
| Neutrophil recruitment | Leukocidin production | Tissue necrosis | Anti-toxin therapy |
| Complement activation | C5a peptidase | Immune evasion | Complement enhancement |
| Antibody production | IgA protease | Mucosal colonization | Passive immunization |
| Macrophage activation | Catalase/SOD | Intracellular survival | Immune stimulation |
| Fever response | Heat shock proteins | Bacterial adaptation | Antipyretic caution |
- Respiratory tract: Ciliary dysfunction + mucus hypersecretion
- Urinary tract: Ascending infection via ureteral reflux
- Bloodstream: Endothelial damage + coagulation cascade activation
- CNS: Blood-brain barrier disruption + cerebral edema
⭐ Clinical Pearl: Bacterial load >10⁵ CFU/mL in normally sterile sites indicates active infection requiring immediate treatment, while <10³ CFU/mL suggests contamination or early colonization in >95% of cases
Emerging Resistance Networks - Evolutionary Adaptation
Bacterial resistance spreads through interconnected networks involving horizontal gene transfer, selective pressure, and environmental reservoirs that create global resistance patterns.
- Horizontal Gene Transfer Mechanisms:
- Conjugation: Plasmid transfer at rates of 10⁻³-10⁻⁵ per donor cell
- Transformation: DNA uptake from environment
- Transduction: Bacteriophage-mediated gene transfer
- Transposition: Mobile genetic elements carrying resistance genes
- Resistance Dissemination Patterns:
- Healthcare settings: Clonal spread of resistant organisms
- Community reservoirs: Commensal bacteria as resistance gene pools
- Agricultural use: Selection pressure from antibiotic use in livestock
- Global travel: Rapid international resistance gene dissemination
💡 Master This: Carbapenem-resistant Enterobacteriaceae (CRE) spread through plasmid-mediated resistance genes that can transfer between species within hours of co-culture, explaining why single CRE cases require immediate contact isolation and molecular surveillance to prevent hospital outbreaks
These pathogenic networks demonstrate how bacterial success depends on coordinated virulence strategies that exploit host vulnerabilities while adapting to therapeutic interventions, revealing targets for next-generation antimicrobial approaches.
🌐 Pathogenic Networks — Multi-System Integration
🎯 Clinical Mastery Arsenal - Rapid Reference Framework
Essential Bacterial Arsenal - Critical Numbers and Thresholds
- Diagnostic Thresholds:
- Blood cultures: ≥1 positive bottle indicates bacteremia
- Urine cultures: ≥10⁵ CFU/mL confirms UTI (≥10² CFU/mL in symptomatic women)
- CSF cultures: Any growth in sterile CSF indicates meningitis
- Respiratory cultures: ≥10⁴ CFU/mL suggests pneumonia in BAL specimens
- Resistance Breakpoints:
- MRSA: Oxacillin MIC ≥4 μg/mL
- ESBL: Ceftazidime MIC ≥2 μg/mL with ≥3-fold reduction with clavulanate
- Vancomycin resistance: MIC ≥16 μg/mL (VRE)
- Carbapenem resistance: Meropenem MIC ≥4 μg/mL
📌 Remember: TIMES for critical culture timing - Time to positivity (<24h suggests high bacterial load), Incubation requirements, Media selection, Environmental conditions, Specimen quality
Rapid Pattern Recognition - Clinical Correlation Matrix
| Clinical Syndrome | Most Likely Organisms | Key Diagnostic Features | Empirical Therapy |
|---|---|---|---|
| Healthcare pneumonia | MRSA, Pseudomonas | Ventilator >48h | Vancomycin + anti-pseudomonal β-lactam |
| Bacterial meningitis | S. pneumoniae, N. meningitidis | CSF WBC >1000 | Ceftriaxone + vancomycin |
| Complicated UTI | E. coli, Klebsiella | Fever + flank pain | Fluoroquinolone or ceftriaxone |
| Skin/soft tissue | S. aureus, Streptococcus | Purulent drainage | Clindamycin or doxycycline |
| Intra-abdominal | E. coli, Bacteroides | Polymicrobial growth | Piperacillin-tazobactam |
- High-Yield Clinical Pearls:
- Gram-positive cocci in clusters from blood = S. aureus until proven otherwise
- Oxidase-positive Gram-negative rods = Pseudomonas or Acinetobacter
- Catalase-negative Gram-positive cocci = Streptococcus or Enterococcus
- Spore-forming Gram-positive rods = Bacillus (aerobic) or Clostridium (anaerobic)
⭐ Clinical Pearl: Positive blood cultures in <12 hours indicate high-grade bacteremia (>10³ CFU/mL) requiring immediate source control and aggressive antimicrobial therapy to prevent septic shock and organ failure
Antimicrobial Stewardship Principles - Precision Prescribing
- De-escalation Strategy:
- 48-72 hours: Narrow spectrum based on culture results
- Biomarker guidance: Procalcitonin <0.25 ng/mL suggests bacterial clearance
- Clinical improvement: Fever resolution + WBC normalization
- Duration Optimization:
- Uncomplicated infections: 5-7 days for most bacterial infections
- Complicated infections: 10-14 days with source control
- Endocarditis: 4-6 weeks depending on organism and valve involvement
- Combination Therapy Indications:
- Severe sepsis: Empirical broad coverage until organism identified
- Resistant organisms: Synergistic combinations for CRE, MRSA
- Endocarditis: β-lactam + aminoglycoside for enterococcal infections
💡 Master This: Antibiotic stewardship reduces resistance development by 30-50%, healthcare costs by 20-30%, and adverse events by 25-40% through systematic de-escalation protocols and duration optimization based on clinical response markers
This clinical mastery framework transforms complex bacteriology into systematic decision-making tools that enable rapid, accurate diagnosis and optimal therapeutic management across all infectious disease scenarios.
🎯 Clinical Mastery Arsenal — Rapid Reference Framework
Practice Questions: Bacteria
Test your understanding with these related questions
A 42-year-old woman comes to her primary care physician with 2 days of fever and malaise. She also says that she has a painful red lesion on her left hand that she noticed after shucking oysters at a recent family reunion. Physical exam reveals a well-demarcated swollen, tender, warm, red lesion on her left hand. Pressing the lesion causes a small amount of purulent drainage. The material is cultured and the causative organism is identified. Which of the following characteristics describes the organism that is most associated with this patient's mechanism of infection?












