RNA splicing and alternative splicing US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for RNA splicing and alternative splicing. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
RNA splicing and alternative splicing US Medical PG Question 1: A 4-year-old boy with beta thalassemia requires regular blood transfusions a few times per month because of persistent anemia. He is scheduled for a splenectomy in the next several months. Samples obtained from the boy’s red blood cells show a malformed protein with a length of 160 amino acids (in normal, healthy red blood cells, the functional protein has a length of 146 amino acids). Which of the following best accounts for these findings?
- A. Nonsense mutation
- B. Silent mutation
- C. Missense mutation
- D. Splice site mutation (Correct Answer)
- E. Frameshift mutation
RNA splicing and alternative splicing Explanation: ***Splice site mutation***
- A **splice site mutation** can lead to the retention of an **intron** or the **skipping of an exon**, resulting in an abnormal mRNA sequence.
- If a cryptic splice site is used or an intron is retained, it can lead to the inclusion of additional amino acids in the final protein, thus increasing its length from 146 to 160 amino acids.
*Nonsense mutation*
- A **nonsense mutation** results in a **premature stop codon**, which would produce a **truncated protein** shorter than 146 amino acids.
- This type of mutation does not explain the observed increase in protein length.
*Silent mutation*
- A **silent mutation** changes a single nucleotide but does **not alter the amino acid sequence** of the protein due to the redundancy of the genetic code.
- This would result in a normal protein length of 146 amino acids and no observed malformation.
*Missense mutation*
- A **missense mutation** changes a single nucleotide leading to a **different amino acid**, but it typically **does not alter the total length** of the protein.
- While it can lead to a *malformed protein*, it wouldn't explain the increased length from 146 to 160 amino acids.
*Frameshift mutation*
- A **frameshift mutation** is caused by the **insertion or deletion of nucleotides** not divisible by three, leading to a shift in the reading frame downstream.
- This often results in a **premature stop codon** and a **shorter, non-functional protein**, or a completely altered sequence that is usually unstable, rather than a longer protein with 160 amino acids.
RNA splicing and alternative splicing US Medical PG Question 2: A 20-year-old female presents to the emergency department with squeezing right upper quadrant pain worse after eating. She has a history of a microcytic, hypochromic anemia with target cells. Physical exam shows severe tenderness to palpation in the right upper quadrant and a positive Murphy's sign. By genetic analysis a single point mutation is detected in the gene of interest. Despite this seemingly minor mutation, the protein encoded by this gene is found to be missing a group of 5 consecutive amino acids though the amino acids on either side of this sequence are preserved. This point mutation is most likely located in which of the following regions of the affected gene?
- A. Intron (Correct Answer)
- B. Exon
- C. Kozak consensus sequence
- D. Transcriptional promoter
- E. Polyadenylation sequence
RNA splicing and alternative splicing Explanation: ***Intron***
- A point mutation at an **intron-exon boundary** (splice donor or acceptor site) can disrupt normal splicing, leading to **exon skipping** during mRNA processing. If the skipped exon encodes exactly 5 amino acids, the resulting protein will be missing this specific sequence while all flanking amino acids remain intact.
- This is the classic mechanism for many **thalassemias**. The clinical presentation of **microcytic, hypochromic anemia with target cells** plus **cholecystitis** (from chronic hemolysis causing pigmented gallstones) strongly suggests a hemoglobinopathy caused by a splice site mutation.
- Splice site mutations (located in introns at exon-intron boundaries) are among the most common causes of beta-thalassemia and can result in precise, predictable deletions of amino acid sequences.
*Exon*
- A point mutation within the coding sequence of an **exon** typically causes a **single amino acid substitution** (missense), a **premature stop codon** (nonsense), or if it's an insertion/deletion, a **frameshift mutation**.
- A point mutation within an exon **cannot** cause the deletion of exactly 5 consecutive amino acids while preserving the flanking sequences. This pattern is characteristic of exon skipping, not intra-exonic mutations.
*Kozak consensus sequence*
- The **Kozak sequence** surrounds the start codon and affects **translation initiation efficiency**. Mutations here would reduce the amount of protein produced but would not cause internal deletions of specific amino acid sequences.
- It does not explain the deletion of 5 consecutive amino acids from the middle of the protein.
*Transcriptional promoter*
- Mutations in the **promoter region** affect the **rate of transcription**, leading to increased or decreased mRNA levels.
- They do not alter the amino acid sequence of the protein or cause specific internal deletions.
*Polyadenylation sequence*
- The **polyadenylation signal** is important for **mRNA stability and 3' end processing**.
- Mutations here affect mRNA stability and abundance but do not change the amino acid sequence or cause internal deletions within the protein.
RNA splicing and alternative splicing US Medical PG Question 3: A 21-year-old woman comes to the physician for an annual health maintenance examination. She has no particular health concerns. Laboratory studies show:
Hemoglobin 11.2 g/dL
Mean corpuscular volume 74 μm3
Mean corpuscular hemoglobin concentration 30% Hb/cell
Red cell distribution width 14% (N=13–15)
Genetic analysis shows a point mutation in intron 1 of a gene on the short arm of chromosome 11. A process involving which of the following components is most likely affected in this patient?
- A. TATA-rich nucleotide sequence
- B. Transfer RNA
- C. Heat shock protein 60
- D. Small nuclear ribonucleoprotein (Correct Answer)
- E. MicroRNA
RNA splicing and alternative splicing Explanation: ***Small nuclear ribonucleoprotein***
- The patient's lab results (low 11.2 g/dL **hemoglobin**, low 74 µm3 **MCV**, and low 30% **MCHC**) indicate **microcytic, hypochromic anemia**, consistent with **thalassemia**.
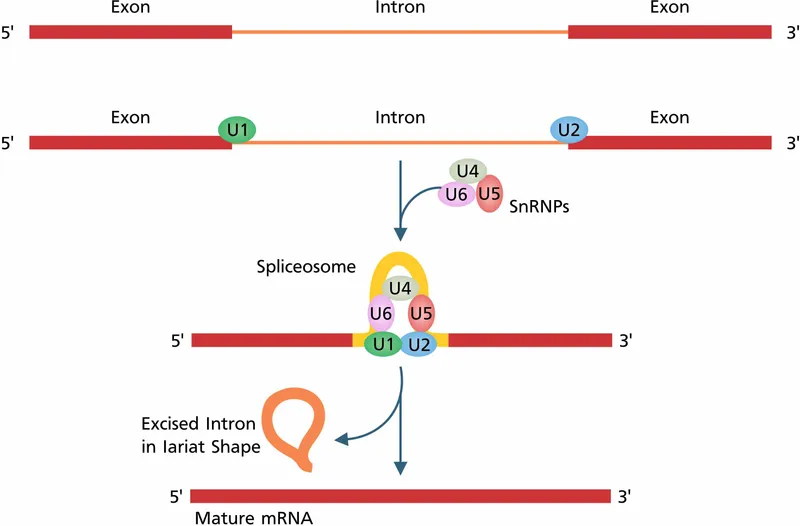
- A point mutation in **intron 1** of a gene suggests a problem with **RNA splicing**, which is mediated by **small nuclear ribonucleoproteins (snRNPs)** as part of the spliceosome.
*TATA-rich nucleotide sequence*
- The **TATA box** is located in the **promoter region** of genes and is involved in the initiation of **transcription**, not splicing.
- A mutation in the TATA box would affect the **rate of transcription** or gene expression, not the processing of mRNA after transcription.
*Transfer RNA*
- **tRNA** molecules are essential for **protein translation** by carrying specific amino acids to the ribosome.
- A problem with tRNA would affect the **synthesis of proteins**, not the processing of pre-mRNA.
*Heat shock protein 60*
- **Heat shock proteins** are molecular **chaperones** involved in the proper **folding of proteins** and preventing protein aggregation.
- A defect in HSP60 would lead to misfolded proteins, not impaired mRNA splicing.
*MicroRNA*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by **silencing mRNA** or inhibiting **translation**.
- While miRNAs regulate gene expression, they are not directly involved in the **splicing of introns** from pre-mRNA.
RNA splicing and alternative splicing US Medical PG Question 4: A 32-year-old woman comes to the physician because of a 4-day history of low-grade fever, joint pain, and muscle aches. The day before the onset of her symptoms, she was severely sunburned on her face and arms during a hike with friends. She also reports being unusually fatigued over the past 3 months. Her only medication is a combined oral contraceptive pill. Her temperature is 37.9°C (100.2°F). Examination shows bilateral swelling and tenderness of the wrists and metacarpophalangeal joints. There are multiple nontender superficial ulcers on the oral mucosa. The detection of antibodies directed against which of the following is most specific for this patient's condition?
- A. Nuclear Sm proteins (Correct Answer)
- B. Fc region of IgG
- C. Single-stranded DNA
- D. Cell nucleus
- E. Histones
RNA splicing and alternative splicing Explanation: ***Nuclear Sm proteins***
- Antibodies to **Sm proteins** (anti-Sm antibodies) are highly specific for **Systemic Lupus Erythematosus (SLE)**, although present in only a minority of patients.
- The patient's symptoms, including **photosensitivity (exacerbation by sunburn)**, **arthritis**, **oral ulcers**, and **fatigue**, are classic manifestations of SLE.
*Fc region of IgG*
- Antibodies directed against the **Fc region of IgG** are known as **rheumatoid factor (RF)**.
- While RF can be positive in a small percentage of SLE patients, it is most characteristic of **rheumatoid arthritis** and is not specific for SLE.
*Single-stranded DNA*
- Antibodies to **single-stranded DNA (anti-ssDNA antibodies)** are found in various autoimmune diseases, including SLE, but are **less specific** than anti-dsDNA or anti-Sm antibodies for SLE diagnosis.
- These antibodies can also be seen in drug-induced lupus and other conditions, making them a less definitive marker.
*Cell nucleus*
- Antibodies directed against the **cell nucleus** (antinuclear antibodies or **ANA**) are present in nearly all patients with SLE and are highly sensitive for the disease.
- However, ANA can also be positive in many other autoimmune conditions, infections, and even healthy individuals, making it **not specific** enough for a definitive diagnosis without other criteria.
*Histones*
- Antibodies to **histones** are most commonly associated with **drug-induced lupus erythematosus**.
- While they can be present in some cases of SLE, the patient's presentation does not strongly suggest drug-induced lupus, and anti-histone antibodies are not the most specific marker for typical SLE.
RNA splicing and alternative splicing US Medical PG Question 5: A 9-year-old boy is getting fitted for leg braces because he has become too weak to walk without them. He developed normally until age 3 but then he began to get tired more easily and fell a lot. Over time he started having trouble walking and would stand up by using the Gower maneuver. Despite this weakness, his neurologic development is normal for his age. On exam his calves appeared enlarged and he was sent for genetic testing. Sequence data showed that he had a mutation causing a shift in the reading frame, resulting in a severely truncated and non-functional protein. Which of the following types of mutations is most likely the cause of this patient's disorder?
- A. Splice site
- B. Missense
- C. Nonsense
- D. Frameshift (Correct Answer)
- E. Silent
RNA splicing and alternative splicing Explanation: ***Frameshift***
- A **frameshift mutation** is caused by the insertion or deletion of nucleotides not in multiples of three, leading to a shift in the reading frame of the mRNA. This results in altered codons downstream of the mutation, typically leading to a **premature stop codon** and a **severely truncated, non-functional protein**.
- The description of a mutation causing "a shift in the reading frame, resulting in a severely truncated and non-functional protein" is characteristic of a frameshift mutation, which is the most common type of mutation in **Duchenne muscular dystrophy** (DMD). The clinical picture (onset around age 3-5, progressive proximal weakness, Gower maneuver, calf pseudohypertrophy) is classic for DMD.
- In DMD, frameshift mutations in the dystrophin gene lead to complete loss of functional dystrophin protein, causing the severe progressive muscle weakness seen in this patient.
*Splice site*
- A **splice site mutation** affects the recognition sequences for intron-exon boundaries during mRNA splicing, potentially leading to exon skipping, intron retention, or use of cryptic splice sites. While splice site mutations can cause DMD (accounting for ~10% of cases), they are less common than frameshifts/deletions.
- The specific description of a "shift in the reading frame" points more directly to a frameshift mutation rather than a splicing defect.
*Missense*
- A **missense mutation** results in a single nucleotide substitution that changes one codon to specify a different amino acid. This produces a full-length protein with a single amino acid substitution.
- Missense mutations typically cause the milder **Becker muscular dystrophy** phenotype (with partially functional dystrophin), not the severe Duchenne phenotype described here. The description of a "severely truncated and non-functional protein" does not fit a missense mutation.
*Nonsense*
- A **nonsense mutation** introduces a premature stop codon directly by changing a codon that normally specifies an amino acid into a stop codon (UAG, UAA, or UGA). This results in a truncated protein.
- While nonsense mutations can cause DMD and do produce truncated proteins, the specific wording "shift in the reading frame" is more characteristic of a frameshift mutation. Nonsense mutations don't shift the reading frame—they directly create a stop signal.
*Silent*
- A **silent (synonymous) mutation** is a nucleotide substitution that does not change the amino acid sequence due to the degeneracy of the genetic code (multiple codons can specify the same amino acid).
- Silent mutations produce normal, full-length proteins and would not cause disease symptoms.
RNA splicing and alternative splicing US Medical PG Question 6: An investigator is studying the crossbridge cycle of muscle contraction. Tissue from the biceps brachii muscle is obtained at the autopsy of an 87-year-old man. Investigation of the muscle tissue shows myosin heads attached to actin filaments. Binding of myosin heads to which of the following elements would most likely cause detachment of myosin from actin filaments?
- A. ATP (Correct Answer)
- B. Troponin C
- C. Tropomyosin
- D. ADP
- E. cGMP
RNA splicing and alternative splicing Explanation: ***ATP***
- The binding of **ATP** to the **myosin head** causes a conformational change that reduces its affinity for actin, leading to detachment.
- This step is crucial for the muscle to relax and for the subsequent power stroke to occur.
*Troponin C*
- **Troponin C** is a regulatory protein that binds calcium, which then causes a conformational change in the troponin-tropomyosin complex, revealing the **actin binding sites** for myosin.
- It does not directly cause myosin detachment; instead, it facilitates the binding of myosin to actin.
*Tropomyosin*
- **Tropomyosin** is a long, fibrous protein that covers the **myosin-binding sites** on actin in a relaxed muscle, preventing cross-bridge formation.
- Its movement, regulated by troponin, allows myosin to bind, but it does not directly cause detachment.
*ADP*
- **ADP** is released from the myosin head during the power stroke, but its binding does not cause detachment; rather, it is present during the strongly bound state before **ATP** binds.
- The presence of **ADP** and inorganic phosphate (Pi) often promotes the strong binding of myosin to actin.
*cGMP*
- **cGMP** (cyclic guanosine monophosphate) is a second messenger involved in various cellular processes, including smooth muscle relaxation, but it is not directly involved in the cross-bridge cycle and detachment of **myosin from actin** in skeletal muscle.
- Its primary role in muscle physiology is often linked to nitric oxide signaling and vasodilation.
RNA splicing and alternative splicing US Medical PG Question 7: A 30-year-old African American woman develops a facial rash in a "butterfly" pattern over her face and complains of feeling tired and achy in her joints. In the course of a full rheumatologic workup you note that she has anti-snRNP antibodies. Which of the following do snRNPs affect?
- A. Transcription of mRNA
- B. Intron removal from the mRNA (Correct Answer)
- C. Protection of mRNA from degradation
- D. Polyadenylation of the 3' end of mRNA
- E. Addition of the 5' 7-methylguanosine cap of mRNA
RNA splicing and alternative splicing Explanation: ***Intron removal from the mRNA***
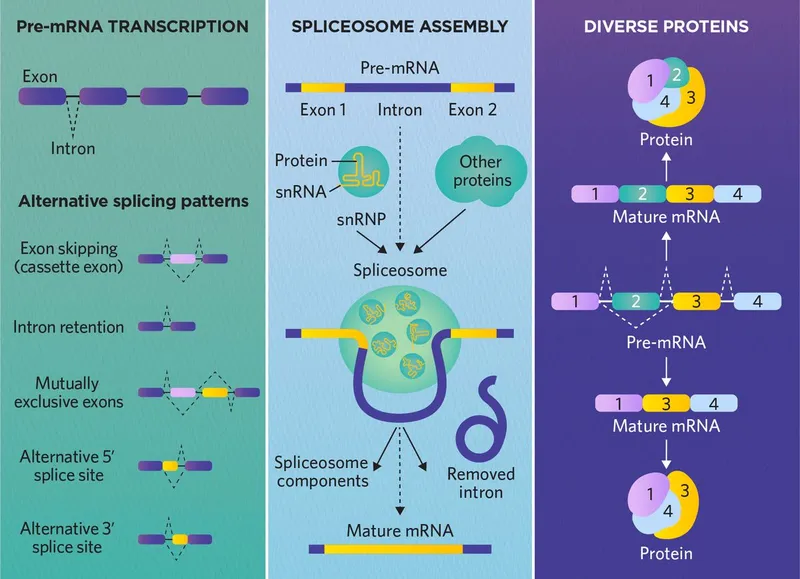
- **Small nuclear ribonucleoproteins (snRNPs)** are crucial components of the **spliceosome**, the molecular machinery responsible for removing non-coding introns from pre-mRNA.
- **snRNPs** recognize and bind to specific sequences within introns and at exon-intron junctions, guiding the splicing process to produce mature mRNA.
*Transcription of mRNA*
- **Transcription** is the process where DNA is copied into RNA, primarily catalyzed by **RNA polymerase**.
- While snRNPs are involved in post-transcriptional modification, they do not directly affect the initial synthesis of the mRNA transcript.
*Protection of mRNA from degradation*
- The **poly-A tail** and the **5' cap** play significant roles in protecting mRNA from degradation by exonucleases.
- While splicing is essential for producing a functional message, snRNPs themselves are not primarily involved in the degradation protection mechanism.
*Polyadenylation of the 3' end of mRNA*
- **Polyadenylation** involves the addition of a **poly-A tail** to the 3' end of the mRNA, which is mediated by poly-A polymerase.
- This process is distinct from splicing and occurs after the mature mRNA has been formed.
*Addition of the 5' 7-methylguanosine cap of mRNA*
- The **5' cap**, a 7-methylguanosine residue, is added to the 5' end of the mRNA during transcription and is crucial for ribosome binding and mRNA stability.
- This capping process occurs early in mRNA synthesis and is not directly mediated by snRNPs.
RNA splicing and alternative splicing US Medical PG Question 8: A 25-year-old female comes to the clinic complaining of fatigue and palpitations. She has been undergoing immense stress from her thesis defense and has been extremely tired. The patient denies any weight loss, diarrhea, cold/heat intolerance. TSH was within normal limits. She reports a family history of "blood disease" and was later confirmed positive for β-thalassemia minor. It is believed that abnormal splicing of the beta globin gene results in β-thalassemia. What is removed during this process that allows RNA to be significantly shorter than DNA?
- A. 3'-poly(A) tail
- B. Exons
- C. Introns (Correct Answer)
- D. microRNAs
- E. snRNPs
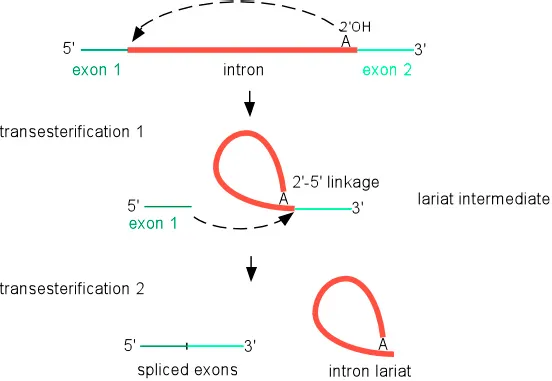
RNA splicing and alternative splicing Explanation: **Introns**
- **Introns** are non-coding regions within a gene that are removed from the pre-mRNA transcript during **splicing**.
- This removal and the subsequent ligation of exons lead to a mature mRNA molecule that is significantly shorter than the initial DNA template.
*3'-poly(A) tail*
- The **3'-poly(A) tail** is an addition to the 3' end of the mRNA molecule, not a removed segment during splicing, and it provides stability and aids in translation.
- While it contributes to mRNA processing, its addition does not involve removing existing sequences to shorten the transcript.
*Exons*
- **Exons** are the coding regions of a gene that are retained and ligated together to form the mature mRNA, which is then translated into protein.
- If exons were removed, the resulting protein would be truncated or non-functional, and the mRNA would not contain the necessary genetic information.
*microRNAs*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by inhibiting translation or promoting mRNA degradation.
- They are not part of the pre-mRNA transcript that is processed into mRNA; rather, they are distinct regulatory molecules.
*snRNPs*
- **Small nuclear ribonucleoproteins (snRNPs)** are components of the spliceosome, the molecular machine responsible for carrying out splicing.
- They are involved in the process of intron removal but are not themselves removed from the RNA; they are catalytic machinery.
RNA splicing and alternative splicing US Medical PG Question 9: A scientist wants to determine if a specific fragment is contained within genome X. She uses a restriction enzyme to digest the genome into smaller fragments to run on an agarose gel, with the goal of separating the resulting fragments. A nitrocellulose blotting paper is then used to transfer the fragments from the agarose gel. A radiolabeled probe containing a complementary sequence to the fragment she is searching for is incubated with the blotting paper. Which of the following is the RNA equivalent of this technique?
- A. RT-PCR
- B. Western blot
- C. qPCR
- D. Northern blot (Correct Answer)
- E. Southern blot
RNA splicing and alternative splicing Explanation: **Northern blot**
- The technique described in the question, involving **restriction enzyme digestion**, **agarose gel electrophoresis**, **blotting onto a membrane**, and **hybridization with a labeled probe**, is characteristic of a **Southern blot** for DNA
- The **Northern blot** is the analogous technique used to detect and quantify **RNA** sequences, following the same principles of separation by size and detection by hybridization with a complementary probe
- Both Southern and Northern blots use the same workflow: separate nucleic acids by size on gel → transfer to membrane → detect with complementary probe
*RT-PCR*
- **Reverse transcriptase polymerase chain reaction (RT-PCR)** is used to amplify specific **RNA** sequences by first converting **RNA** into **complementary DNA (cDNA)** using reverse transcriptase, followed by standard PCR
- Unlike Northern blot, it is an **amplification technique** rather than a direct visualization method via blotting
*Western blot*
- **Western blot** is a technique used to detect and identify specific **proteins**, not nucleic acids
- It involves **gel electrophoresis** to separate proteins by size, followed by transfer to a membrane and detection using **antibodies** rather than nucleic acid probes
*qPCR*
- **Quantitative polymerase chain reaction (qPCR)**, also known as real-time PCR, is a technique used to **quantify DNA or RNA** (after reverse transcription) in real-time
- It measures the accumulation of fluorescent signal during the PCR reaction, allowing for real-time monitoring and quantification, which is fundamentally different from a blotting technique
*Southern blot*
- The description in the question *is* a **Southern blot**, which is used for **DNA** detection, not RNA
- Since the question asks for the **RNA equivalent** of the described technique, and Southern blot detects DNA, Northern blot is the correct answer
RNA splicing and alternative splicing US Medical PG Question 10: A 15-year-old boy presents with shortness of breath on exertion for the past 2 weeks. Although he does not have any other complaints, he is concerned about not gaining much weight despite a good appetite. His height is 188 cm (6 ft 2 in) and weight is 58 kg (124 lb). His blood pressure is 134/56 mm Hg and his pulse rate is 78/min. On cardiac auscultation, his apex beat is displaced laterally with a diastolic murmur lateral to the left sternal border. Slit-lamp examination shows an upward and outward displacement of both lenses. Synthesis of which of the following proteins is most likely defective in this patient?
- A. Fibronectin
- B. Elastin
- C. Fibrillin (Correct Answer)
- D. Reticular fibers
- E. Laminin
RNA splicing and alternative splicing Explanation: ***Fibrillin***
- The patient's presentation with **tall stature**, **arachnodactyly** (implied by tall, thin build), **ectopia lentis** (upward and outward lens displacement), and a **diastolic murmur** (suggesting aortic root dilation or dissection, or mitral valve prolapse) are classic features of **Marfan syndrome**.
- **Marfan syndrome** is caused by a defect in the gene encoding **fibrillin-1**, a glycoprotein essential for the formation of elastic fibers and connective tissue integrity.
*Fibronectin*
- **Fibronectin** is involved in cell adhesion, growth, migration, and differentiation, and plays a crucial role in wound healing and embryonic development.
- While essential for connective tissue, defects in fibronectin are not typically associated with the constellation of symptoms seen in Marfan syndrome.
*Elastin*
- **Elastin** works in conjunction with fibrillin to provide elasticity to tissues like the skin, lungs, and blood vessels.
- While Marfan syndrome affects elastic fibers, the primary defect is in fibrillin, which then impairs the proper formation and function of elastin-containing microfibrils.
*Reticular fibers*
- **Reticular fibers** are fine collagen fibers (primarily type III collagen) that form a delicate supporting network in various tissues and organs.
- Defects in reticular fibers are not characteristic of Marfan syndrome; Marfan syndrome is specifically linked to fibrillin defects.
*Laminin*
- **Laminins** are major proteins of the **basal lamina**, essential for cell adhesion and differentiation in epithelial and endothelial tissues.
- Genetic defects in laminin components are often associated with muscular dystrophies or epidermolysis bullosa, not the Marfanoid features presented.
More RNA splicing and alternative splicing US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.