Regulation of gene expression US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Regulation of gene expression. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
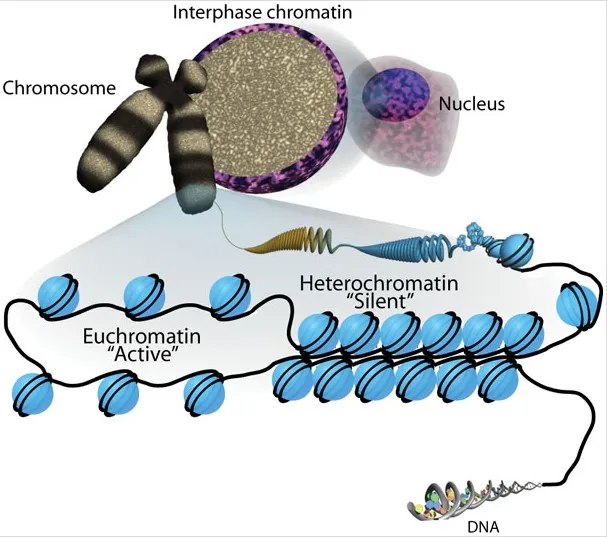
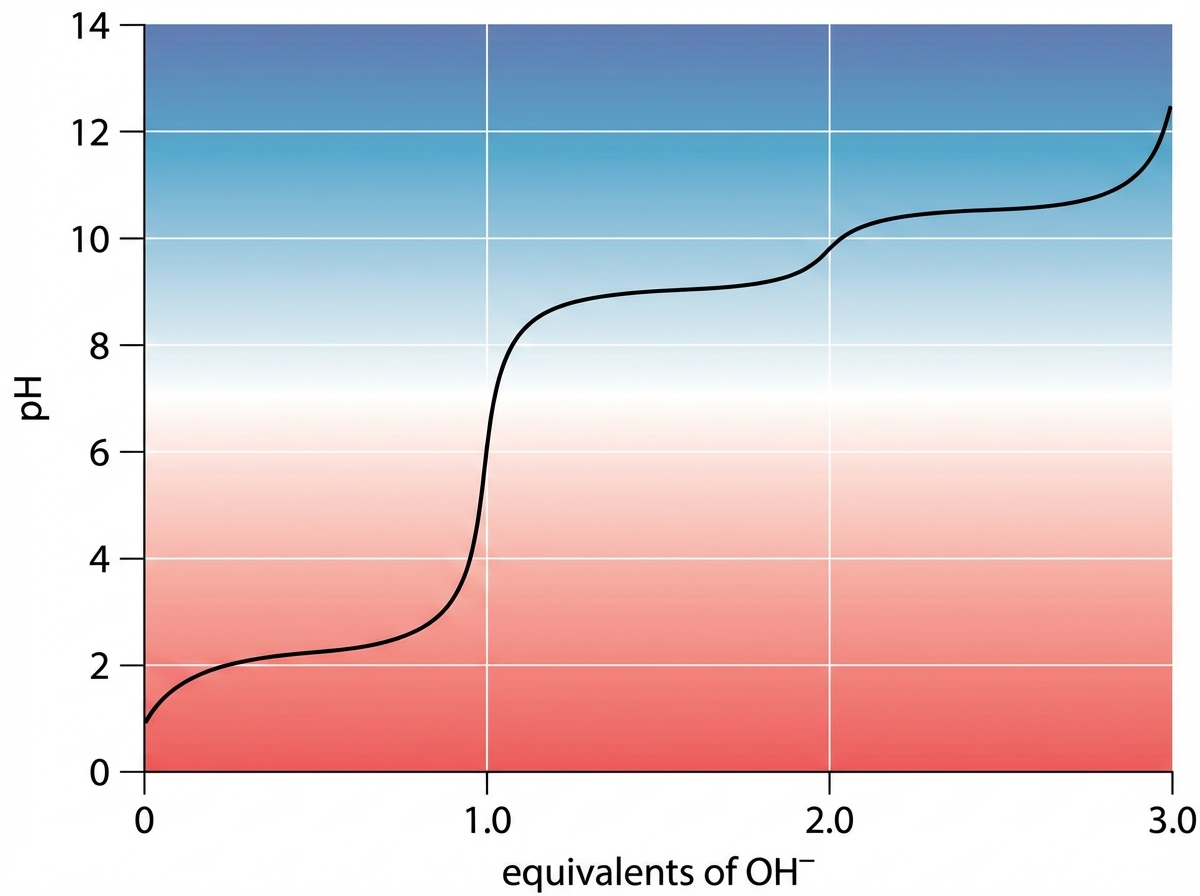
Regulation of gene expression US Medical PG Question 1: An investigator studying epigenetic mechanisms isolates histone proteins, the structural motifs involved in DNA binding and regulation of transcription. The peptide bonds of histone proteins are hydrolyzed and one type of amino acid is isolated. At normal body pH, this amino acid has a net charge of +1 . The investigator performs titration of this amino acid and obtains the graph shown. The isolated amino acid is most likely which of the following?
- A. Proline
- B. Lysine (Correct Answer)
- C. Aspartate
- D. Serine
- E. Histidine
Regulation of gene expression Explanation: ***Lysine***
- Histones are **positively charged** proteins rich in **basic amino acids** like lysine and arginine, which allows them to bind tightly to the negatively charged DNA.
- The titration curve shown with three distinct pKa values and a net charge of +1 at normal body pH (around 7.4) is characteristic of **lysine**, which has both an alpha-amino group (pKa ~9-10) and a basic side chain (pKa ~10.5).
*Proline*
- **Proline is a nonpolar** amino acid that does not contribute significantly to the positive charge of histones required for DNA binding.
- Furthermore, its unique cyclic structure incorporates its amino group into the ring, impacting its pKa relative to other primary amino acids but not making it a primary basic residue in the histone context.
*Aspartate*
- **Aspartate is an acidic amino acid** with a negatively charged side chain at physiological pH, which would repel DNA rather than bind to it.
- Its titration curve would show a net negative charge at normal body pH, not a positive one.
*Serine*
- **Serine is a polar, uncharged** amino acid and would not contribute the necessary positive charge for histone-DNA interaction.
- Its side chain lacks an ionizable group within the physiological pH range, so its titration curve would only show two pKa values (for the carboxyl and amino groups) and a net charge of 0 at neutral pH.
*Histidine*
- While **histidine is a basic amino acid**, its side chain pKa is around 6.0, meaning it is only partially protonated and positively charged at physiological pH.
- A protein rich in **histidine** would not consistently carry a strong positive charge across the typical physiological pH range as effectively as one rich in lysine or arginine.
Regulation of gene expression US Medical PG Question 2: E. coli has the ability to regulate its enzymes to break down various sources of energy when available. It prevents waste by the use of the lac operon, which encodes a polycistronic transcript. At a low concentration of glucose and absence of lactose, which of the following occurs?
- A. Decreased cAMP levels result in poor binding to the catabolite activator protein
- B. Increased cAMP levels result in binding to the catabolite activator protein (Correct Answer)
- C. Increased allolactose levels bind to the repressor
- D. Repressor releases from lac operator
- E. Transcription of the lac Z, Y, and A genes increase
Regulation of gene expression Explanation: ***Increased cAMP levels result in binding to the catabolite activator protein***
- In the absence of glucose, **adenylate cyclase** activity increases, leading to higher levels of **cAMP**.
- **cAMP** then binds to the **catabolite activator protein (CAP)**, forming the **cAMP-CAP complex**, which is crucial for activating lac operon transcription in the absence of glucose.
*Decreased cAMP levels result in poor binding to the catabolite activator protein*
- **Decreased glucose levels** actually lead to **increased cAMP** synthesis, not decreased.
- High **cAMP** levels enhance, not hinder, its binding to **CAP**.
*Increased allolactose levels bind to the repressor*
- **Allolactose** is an inducer that forms in the presence of **lactose**, which is stated to be absent in this scenario.
- Therefore, **allolactose levels** would be low, and it would not bind to the **repressor**.
*Repressor releases from lac operator*
- The **repressor protein** is bound to the **lac operator** in the absence of lactose.
- For the **repressor to be released**, **allolactose** (formed from lactose) must be present to bind to it.
*Transcription of the lac Z, Y, and A genes increase*
- While **cAMP-CAP binding** would promote transcription, the **absence of lactose** means the **repressor remains bound** to the operator.
- This binding effectively blocks RNA polymerase, preventing significant transcription of the **lac Z, Y, and A genes**, regardless of high **cAMP** levels.
Regulation of gene expression US Medical PG Question 3: A 62-year-old woman presents to her primary care physician because of fever, fatigue, and shortness of breath. She has noticed that she has a number of bruises, but she attributes this to a hike she went on 1 week ago. She has diabetes and hypertension well controlled on medication and previously had an abdominal surgery but doesn’t remember why. On physical exam, she has some lumps in her neck and a palpable liver edge. Peripheral blood smear shows white blood cells with peroxidase positive eosinophilic cytoplasmic inclusions. The abnormal protein most likely seen in this disease normally has which of the following functions?
- A. Binding to anti-apoptotic factors
- B. Binding as cofactor to kinases
- C. Inhibiting pro-apoptotic factors
- D. Recruiting histone acetylase proteins (Correct Answer)
- E. Interacting with IL-3 receptor
Regulation of gene expression Explanation: ***Recruiting histone acetylase proteins***
- The clinical presentation with **fever, fatigue, shortness of breath, bruising**, and **hepatomegaly** along with a **peripheral blood smear showing white blood cells with peroxidase-positive eosinophilic cytoplasmic inclusions** strongly suggests **Acute Promyelocytic Leukemia (APL)**.
- APL is characterized by a **chromosomal translocation t(15;17)**, which fuses the **PML (promyelocytic leukemia)** gene with the **RARα (retinoic acid receptor alpha)** gene. The **PML-RARα fusion protein** acts as an **abnormal transcriptional repressor**, recruiting histone deacetylase (HDAC) proteins and forming a complex that blocks gene expression essential for myeloid differentiation, while the **normal RARα protein** (which is being asked about in the question "abnormal protein most likely seen in this disease normally has which of the following functions") normally **recruits histone acetylase proteins** to promote gene transcription crucial for cellular differentiation.
*Binding to anti-apoptotic factors*
- While a protein's function might involve apoptosis regulation, the **PML-RARα fusion protein**'s primary mechanism in APL is related to **transcriptional repression** and **differentiation block**, not directly binding to anti-apoptotic factors in its normal state.
- The normal function of **RARα** is to promote differentiation, and its dysregulation in APL leads to apoptosis resistance due to the block in differentiation and subsequent proliferation, rather than directly binding anti-apoptotic factors.
*Binding as cofactor to kinases*
- **RARα** is a nuclear receptor and a **ligand-activated transcription factor**, not typically known for directly binding as a cofactor to kinases in its normal physiological role.
- Its main function involves **DNA binding** and interaction with co-activator/co-repressor complexes to regulate gene expression.
*Inhibiting pro-apoptotic factors*
- The normal **RARα protein** is involved in promoting cellular differentiation, and its dysregulation in APL leads to a block in differentiation, which can contribute to the accumulation of immature cells that resist apoptosis.
- However, its normal function is not described as directly **inhibiting pro-apoptotic factors**; rather, its absence or altered function in APL leads to an environment that less efficiently triggers apoptosis in undifferentiated cells.
*Interacting with IL-3 receptor*
- The **IL-3 receptor** is a cell surface receptor involved in cytokine signaling that promotes the growth and differentiation of hematopoietic cells.
- **RARα** is a nuclear receptor located within the cell and primarily interacts with **DNA** and **transcriptional co-regulators**, not with cell surface receptors like the IL-3 receptor.
Regulation of gene expression US Medical PG Question 4: A 21-year-old woman comes to the physician for an annual health maintenance examination. She has no particular health concerns. Laboratory studies show:
Hemoglobin 11.2 g/dL
Mean corpuscular volume 74 μm3
Mean corpuscular hemoglobin concentration 30% Hb/cell
Red cell distribution width 14% (N=13–15)
Genetic analysis shows a point mutation in intron 1 of a gene on the short arm of chromosome 11. A process involving which of the following components is most likely affected in this patient?
- A. TATA-rich nucleotide sequence
- B. Transfer RNA
- C. Heat shock protein 60
- D. Small nuclear ribonucleoprotein (Correct Answer)
- E. MicroRNA
Regulation of gene expression Explanation: ***Small nuclear ribonucleoprotein***
- The patient's lab results (low 11.2 g/dL **hemoglobin**, low 74 µm3 **MCV**, and low 30% **MCHC**) indicate **microcytic, hypochromic anemia**, consistent with **thalassemia**.
- A point mutation in **intron 1** of a gene suggests a problem with **RNA splicing**, which is mediated by **small nuclear ribonucleoproteins (snRNPs)** as part of the spliceosome.
*TATA-rich nucleotide sequence*
- The **TATA box** is located in the **promoter region** of genes and is involved in the initiation of **transcription**, not splicing.
- A mutation in the TATA box would affect the **rate of transcription** or gene expression, not the processing of mRNA after transcription.
*Transfer RNA*
- **tRNA** molecules are essential for **protein translation** by carrying specific amino acids to the ribosome.
- A problem with tRNA would affect the **synthesis of proteins**, not the processing of pre-mRNA.
*Heat shock protein 60*
- **Heat shock proteins** are molecular **chaperones** involved in the proper **folding of proteins** and preventing protein aggregation.
- A defect in HSP60 would lead to misfolded proteins, not impaired mRNA splicing.
*MicroRNA*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by **silencing mRNA** or inhibiting **translation**.
- While miRNAs regulate gene expression, they are not directly involved in the **splicing of introns** from pre-mRNA.
Regulation of gene expression US Medical PG Question 5: A scientist is studying the mechanisms by which bacteria become resistant to antibiotics. She begins by obtaining a culture of vancomycin-resistant Enterococcus faecalis and conducts replicate plating experiments. In these experiments, colonies are inoculated onto a membrane and smeared on 2 separate plates, 1 containing vancomycin and the other with no antibiotics. She finds that all of the bacterial colonies are vancomycin resistant because they grow on both plates. She then maintains the bacteria in liquid culture without vancomycin while she performs her other studies. Fifteen generations of bacteria later, she conducts replicate plating experiments again and finds that 20% of the colonies are now sensitive to vancomycin. Which of the following mechanisms is the most likely explanation for why these colonies have become vancomycin sensitive?
- A. Point mutation
- B. Gain of function mutation
- C. Viral infection
- D. Plasmid loss (Correct Answer)
- E. Loss of function mutation
Regulation of gene expression Explanation: ***Plasmid loss***
- The initial **vancomycin resistance** in *Enterococcus faecalis* is often mediated by genes located on **plasmids**, which are extrachromosomal DNA.
- In the absence of selective pressure (vancomycin), bacteria that lose the plasmid (and thus the resistance genes) have a **growth advantage** over those that retain the energetically costly plasmid, leading to an increase in sensitive colonies over generations.
*Point mutation*
- A **point mutation** typically involves a change in a single nucleotide and could lead to loss of resistance if it occurred in a gene conferring resistance.
- However, since there was no selective pressure for loss of resistance, it is less likely that 20% of the population would acquire such a specific point mutation to revert resistance.
*Gain of function mutation*
- A **gain of function mutation** would imply that the bacteria acquired a *new* advantageous trait, not the *loss* of resistance.
- This type of mutation would not explain why some colonies became sensitive to vancomycin after the drug was removed.
*Viral infection*
- **Viral infection** (bacteriophages) can transfer genes through transduction or cause bacterial lysis, but it's not the primary mechanism for a widespread reversion of resistance in the absence of antibiotic pressure.
- It would not explain the observed increase in vancomycin-sensitive colonies due to evolutionary pressure.
*Loss of function mutation*
- While a **loss of function mutation** in a gene conferring resistance could lead to sensitivity, it's generally less likely to explain a 20% shift without selective pressure than **plasmid loss**.
- Plasmids are often unstable and are easily lost in the absence of selection, whereas a specific gene mutation causing loss of function would need to arise and become prevalent in the population.
Regulation of gene expression US Medical PG Question 6: An investigator is studying the effects of zinc deprivation on cancer cell proliferation. It is hypothesized that because zinc is known to be a component of transcription factor motifs, zinc deprivation will result in slower tumor growth. To test this hypothesis, tumor cells are cultured on media containing low and high concentrations of zinc. During the experiment, a labeled oligonucleotide probe is used to identify the presence of a known transcription factor. The investigator most likely used which of the following laboratory techniques?
- A. ELISA
- B. PCR
- C. Western blot
- D. Northern blot
- E. Southwestern blot (Correct Answer)
Regulation of gene expression Explanation: ***Southwestern blot***
- A **Southwestern blot** specifically identifies **DNA-binding proteins** (such as transcription factors) by detecting their ability to bind to specific **labeled DNA oligonucleotide probes**
- The technique involves: protein separation by gel electrophoresis → transfer to membrane → probing with **labeled double-stranded DNA oligonucleotide**
- This directly answers the question: using a labeled oligonucleotide probe to identify a transcription factor
*ELISA*
- **ELISA** detects and quantifies proteins using **antibody-antigen interactions**, not DNA-binding activity
- While it could detect the presence of a transcription factor protein, it cannot assess the protein's ability to bind to specific DNA sequences
- Does not utilize oligonucleotide probes for detection
*PCR*
- **PCR** amplifies specific **DNA sequences** but does not detect or characterize proteins
- This technique would amplify DNA, not identify DNA-binding proteins
- Not applicable for detecting transcription factor presence or function
*Western blot*
- **Western blot** detects specific proteins using **antibodies**, not oligonucleotide probes
- While it could confirm transcription factor protein presence, it cannot assess DNA-binding capability
- Uses antibody-based detection, not nucleotide probe-based detection
*Northern blot*
- **Northern blot** detects specific **RNA molecules**, not DNA-binding proteins
- Uses labeled DNA or RNA probes to detect RNA, not to detect proteins that bind DNA
- Wrong target molecule (RNA vs. proteins)
Regulation of gene expression US Medical PG Question 7: A 28-year-old male presents to his primary care physician with complaints of intermittent abdominal pain and alternating bouts of constipation and diarrhea. His medical chart is not significant for any past medical problems or prior surgeries. He is not prescribed any current medications. Which of the following questions would be the most useful next question in eliciting further history from this patient?
- A. "Does the diarrhea typically precede the constipation, or vice-versa?"
- B. "Is the diarrhea foul-smelling?"
- C. "Please rate your abdominal pain on a scale of 1-10, with 10 being the worst pain of your life"
- D. "Are the symptoms worse in the morning or at night?"
- E. "Can you tell me more about the symptoms you have been experiencing?" (Correct Answer)
Regulation of gene expression Explanation: ***Can you tell me more about the symptoms you have been experiencing?***
- This **open-ended question** encourages the patient to provide a **comprehensive narrative** of their symptoms, including details about onset, frequency, duration, alleviating/aggravating factors, and associated symptoms, which is crucial for diagnosis.
- In a patient presenting with vague, intermittent symptoms like alternating constipation and diarrhea, allowing them to elaborate freely can reveal important clues that might not be captured by more targeted questions.
*Does the diarrhea typically precede the constipation, or vice-versa?*
- While knowing the sequence of symptoms can be helpful in understanding the **pattern of bowel dysfunction**, it is a very specific question that might overlook other important aspects of the patient's experience.
- It prematurely narrows the focus without first obtaining a broad understanding of the patient's overall symptomatic picture.
*Is the diarrhea foul-smelling?*
- Foul-smelling diarrhea can indicate **malabsorption** or **bacterial overgrowth**, which are important to consider in some gastrointestinal conditions.
- However, this is a **specific symptom inquiry** that should follow a more general exploration of the patient's symptoms, as it may not be relevant if other crucial details are missed.
*Please rate your abdominal pain on a scale of 1-10, with 10 being the worst pain of your life*
- Quantifying pain intensity is useful for assessing the **severity of discomfort** and monitoring changes over time.
- However, for a patient with intermittent rather than acute, severe pain, understanding the **character, location, and triggers** of the pain is often more diagnostically valuable than just a numerical rating initially.
*Are the symptoms worse in the morning or at night?*
- Diurnal variation can be relevant in certain conditions, such as inflammatory bowel diseases where nocturnal symptoms might be more concerning, or functional disorders whose symptoms might be stress-related.
- This is another **specific question** that should come after gathering a more complete initial picture of the patient's symptoms to ensure no key information is overlooked.
Regulation of gene expression US Medical PG Question 8: An investigator is studying the effect of chromatin structure on gene regulation. The investigator isolates a class of proteins that compact DNA by serving as spools upon which DNA winds around. These proteins are most likely rich in which of the following compounds?
- A. Phosphate
- B. Disulfide-bonded cysteine
- C. Lysine and arginine (Correct Answer)
- D. Heparan sulfate
- E. Proline and alanine
Regulation of gene expression Explanation: ***Lysine and arginine***
- DNA is **negatively charged** due to its phosphate backbone. Proteins that compact DNA (like **histones**) must be **positively charged** to electrostatically interact with and bind to DNA.
- **Lysine** and **arginine** are positively charged amino acids that are abundant in histones, facilitating this interaction.
*Phosphate*
- **Phosphate** groups are negatively charged and are a major component of the **DNA backbone** itself, not the proteins that compact DNA.
- Proteins rich in phosphate would be negatively charged, which would inhibit DNA binding due to **electrostatic repulsion**.
*Disulfide-bonded cysteine*
- **Cysteine residues** can form disulfide bonds, which are important for maintaining the **tertiary and quaternary structure** of many proteins.
- However, disulfide bonds do not primarily contribute to the basicity or positive charge required for DNA binding; rather, they play a crucial role in protein **folding and stability**.
*Heparan sulfate*
- **Heparan sulfate** is a **glycosaminoglycan** that is negatively charged and found on cell surfaces and in the extracellular matrix.
- It plays roles in cell signaling and adhesion but is not a component of the core histone proteins that compact DNA.
*Proline and alanine*
- **Proline** and **alanine** are common amino acids, but they are **nonpolar** or **neutral** at physiological pH.
- They do not contribute a significant **positive charge** to proteins, which is essential for binding to the negatively charged DNA.
Regulation of gene expression US Medical PG Question 9: A 25-year-old female comes to the clinic complaining of fatigue and palpitations. She has been undergoing immense stress from her thesis defense and has been extremely tired. The patient denies any weight loss, diarrhea, cold/heat intolerance. TSH was within normal limits. She reports a family history of "blood disease" and was later confirmed positive for β-thalassemia minor. It is believed that abnormal splicing of the beta globin gene results in β-thalassemia. What is removed during this process that allows RNA to be significantly shorter than DNA?
- A. 3'-poly(A) tail
- B. Exons
- C. Introns (Correct Answer)
- D. microRNAs
- E. snRNPs
Regulation of gene expression Explanation: **Introns**
- **Introns** are non-coding regions within a gene that are removed from the pre-mRNA transcript during **splicing**.
- This removal and the subsequent ligation of exons lead to a mature mRNA molecule that is significantly shorter than the initial DNA template.
*3'-poly(A) tail*
- The **3'-poly(A) tail** is an addition to the 3' end of the mRNA molecule, not a removed segment during splicing, and it provides stability and aids in translation.
- While it contributes to mRNA processing, its addition does not involve removing existing sequences to shorten the transcript.
*Exons*
- **Exons** are the coding regions of a gene that are retained and ligated together to form the mature mRNA, which is then translated into protein.
- If exons were removed, the resulting protein would be truncated or non-functional, and the mRNA would not contain the necessary genetic information.
*microRNAs*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by inhibiting translation or promoting mRNA degradation.
- They are not part of the pre-mRNA transcript that is processed into mRNA; rather, they are distinct regulatory molecules.
*snRNPs*
- **Small nuclear ribonucleoproteins (snRNPs)** are components of the spliceosome, the molecular machine responsible for carrying out splicing.
- They are involved in the process of intron removal but are not themselves removed from the RNA; they are catalytic machinery.
Regulation of gene expression US Medical PG Question 10: A 30-year-old African American woman develops a facial rash in a "butterfly" pattern over her face and complains of feeling tired and achy in her joints. In the course of a full rheumatologic workup you note that she has anti-snRNP antibodies. Which of the following do snRNPs affect?
- A. Transcription of mRNA
- B. Intron removal from the mRNA (Correct Answer)
- C. Protection of mRNA from degradation
- D. Polyadenylation of the 3' end of mRNA
- E. Addition of the 5' 7-methylguanosine cap of mRNA
Regulation of gene expression Explanation: ***Intron removal from the mRNA***
- **Small nuclear ribonucleoproteins (snRNPs)** are crucial components of the **spliceosome**, the molecular machinery responsible for removing non-coding introns from pre-mRNA.
- **snRNPs** recognize and bind to specific sequences within introns and at exon-intron junctions, guiding the splicing process to produce mature mRNA.
*Transcription of mRNA*
- **Transcription** is the process where DNA is copied into RNA, primarily catalyzed by **RNA polymerase**.
- While snRNPs are involved in post-transcriptional modification, they do not directly affect the initial synthesis of the mRNA transcript.
*Protection of mRNA from degradation*
- The **poly-A tail** and the **5' cap** play significant roles in protecting mRNA from degradation by exonucleases.
- While splicing is essential for producing a functional message, snRNPs themselves are not primarily involved in the degradation protection mechanism.
*Polyadenylation of the 3' end of mRNA*
- **Polyadenylation** involves the addition of a **poly-A tail** to the 3' end of the mRNA, which is mediated by poly-A polymerase.
- This process is distinct from splicing and occurs after the mature mRNA has been formed.
*Addition of the 5' 7-methylguanosine cap of mRNA*
- The **5' cap**, a 7-methylguanosine residue, is added to the 5' end of the mRNA during transcription and is crucial for ribosome binding and mRNA stability.
- This capping process occurs early in mRNA synthesis and is not directly mediated by snRNPs.
More Regulation of gene expression US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.