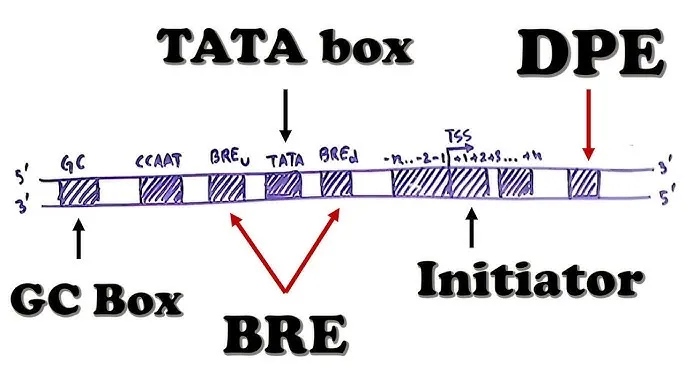
Promoters and transcription factors US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Promoters and transcription factors. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Promoters and transcription factors US Medical PG Question 1: A 33-year-old woman comes to the physician 1 week after noticing a lump in her right breast. Fifteen years ago, she was diagnosed with osteosarcoma of her left distal femur. Her father died of an adrenocortical carcinoma at the age of 41 years. Examination shows a 2-cm, firm, immobile mass in the lower outer quadrant of the right breast. A core needle biopsy of the mass shows adenocarcinoma. Genetic analysis in this patient is most likely to show a defect in which of the following genes?
- A. BRCA1
- B. KRAS
- C. TP53 (Correct Answer)
- D. Rb
- E. PTEN
Promoters and transcription factors Explanation: ***TP53***
- This patient's presentation with **early-onset breast cancer**, a history of **osteosarcoma** at a young age, and a father's death from **adrenocortical carcinoma** at 41 years strongly suggests **Li-Fraumeni syndrome**.
- Li-Fraumeni syndrome is an autosomal dominant disorder caused by a germline mutation in the **tumor suppressor gene TP53**, increasing the risk for multiple primary cancers at a young age.
*BRCA1*
- While **BRCA1 mutations** are associated with an increased risk of breast and ovarian cancer, they are not typically linked to osteosarcoma or adrenocortical carcinoma.
- The constellation of cancers in this patient is more indicative of Li-Fraumeni syndrome than solely a BRCA1-related cancer syndrome.
*KRAS*
- **KRAS** is an oncogene commonly mutated in several cancers, including pancreatic, colorectal, and lung cancer, but is not primarily associated with either Li-Fraumeni syndrome or the specific tumors seen in this family history.
- Mutations in KRAS are typically somatic mutations acquired during a person's lifetime, not germline mutations causing inherited cancer syndromes like the one suggested here.
*Rb*
- Mutations in the **retinoblastoma (Rb) gene** are associated with retinoblastoma and an increased risk of osteosarcoma, but not typically with adrenocortical carcinoma or breast cancer as part of a classic inherited syndrome.
- The combination of breast cancer, osteosarcoma, and adrenocortical carcinoma points more specifically to TP53.
*PTEN*
- **PTEN mutations** are associated with Cowden syndrome, which increases the risk for breast cancer, thyroid cancer, and endometrial cancer, along with benign growths.
- However, Cowden syndrome does not typically include osteosarcoma or adrenocortical carcinoma as prominent features, making PTEN less likely than TP53 for this specific family history.
Promoters and transcription factors US Medical PG Question 2: A 25-year-old female comes to the clinic complaining of fatigue and palpitations. She has been undergoing immense stress from her thesis defense and has been extremely tired. The patient denies any weight loss, diarrhea, cold/heat intolerance. TSH was within normal limits. She reports a family history of "blood disease" and was later confirmed positive for β-thalassemia minor. It is believed that abnormal splicing of the beta globin gene results in β-thalassemia. What is removed during this process that allows RNA to be significantly shorter than DNA?
- A. 3'-poly(A) tail
- B. Exons
- C. Introns (Correct Answer)
- D. microRNAs
- E. snRNPs
Promoters and transcription factors Explanation: **Introns**
- **Introns** are non-coding regions within a gene that are removed from the pre-mRNA transcript during **splicing**.
- This removal and the subsequent ligation of exons lead to a mature mRNA molecule that is significantly shorter than the initial DNA template.
*3'-poly(A) tail*
- The **3'-poly(A) tail** is an addition to the 3' end of the mRNA molecule, not a removed segment during splicing, and it provides stability and aids in translation.
- While it contributes to mRNA processing, its addition does not involve removing existing sequences to shorten the transcript.
*Exons*
- **Exons** are the coding regions of a gene that are retained and ligated together to form the mature mRNA, which is then translated into protein.
- If exons were removed, the resulting protein would be truncated or non-functional, and the mRNA would not contain the necessary genetic information.
*microRNAs*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by inhibiting translation or promoting mRNA degradation.
- They are not part of the pre-mRNA transcript that is processed into mRNA; rather, they are distinct regulatory molecules.
*snRNPs*
- **Small nuclear ribonucleoproteins (snRNPs)** are components of the spliceosome, the molecular machine responsible for carrying out splicing.
- They are involved in the process of intron removal but are not themselves removed from the RNA; they are catalytic machinery.
Promoters and transcription factors US Medical PG Question 3: An investigator studying protein synthesis in human stem cells isolates tRNA molecules bound to mRNA molecules. The isolated tRNA molecules have inosine in the 5' position of the anticodon; of these, some are bound to adenine, some to cytosine, and some to uracil at the 3' position of the mRNA codon. Which of the following properties of the genetic code is best illustrated by this finding?
- A. Unambiguity
- B. Non-overlapping
- C. Degeneracy (Correct Answer)
- D. Specificity of the start codon
- E. Specificity of stop codons
Promoters and transcription factors Explanation: ***Degeneracy***
- The finding that a single tRNA anticodon (with **inosine** at the 5' position) can bind to multiple different mRNA codons (ending in **adenine, cytosine, or uracil**) illustrates the concept of **degeneracy** in the genetic code.
- This **wobble hypothesis** allows fewer tRNAs to recognize more than one codon for a given amino acid, meaning multiple codons can code for the same amino acid.
*Unambiguity*
- The genetic code is unambiguous, meaning that each codon specifies **only one specific amino acid** (or a stop signal) and never two different amino acids.
- This finding, however, shows one tRNA recognizing multiple codons, not one codon coding for multiple amino acids.
*Non-overlapping*
- The **non-overlapping** nature of the genetic code means that each nucleotide in an mRNA sequence is read only once as part of a single codon, without sharing nucleotides between adjacent codons.
- This concept describes how codons are read sequentially, not the flexibility of codon-anticodon pairing.
*Specificity of the start codon*
- The **start codon (AUG)** specifically initiates translation, coding for methionine, and signals the beginning of a polypeptide chain.
- This finding relates to the wobble pairing at the 3' end of the codon, not the initiation of translation.
*Specificity of stop codons*
- **Stop codons (UAA, UAG, UGA)** specifically signal the termination of translation without coding for any amino acid.
- This finding describes the flexibility of codon-anticodon pairing, not the distinct function of termination codons.
Promoters and transcription factors US Medical PG Question 4: A clinical trial is being run with patients that have a genetic condition characterized by abnormal hemoglobin that can undergo polymerization when exposed to hypoxia, acidosis, or dehydration. This process of polymerization is responsible for the distortion of the red blood cell (RBC) that acquires a crescent shape and the hemolysis of RBCs. Researchers are studying the mechanisms of the complications commonly observed in these patients such as stroke, aplastic crisis, and auto-splenectomy. What kind of mutation leads to the development of the disease?
- A. Silent mutation
- B. Splice site
- C. Missense mutation (Correct Answer)
- D. Nonsense mutation
- E. Frameshift mutation
Promoters and transcription factors Explanation: ***Missense mutation***
- A missense mutation results in a **single nucleotide substitution** that changes the codon to code for a different amino acid, altering the protein.
- In **sickle cell disease**, a missense mutation in the beta-globin gene (GAG to GTG) leads to the substitution of **glutamic acid for valine**, causing abnormal hemoglobin (HbS) that polymerizes under deoxygenated conditions.
*Silent mutation*
- A silent mutation is a **point mutation** that results in a new codon that still codes for the **same amino acid**, meaning there is no change in the protein sequence.
- Therefore, it would not lead to an **abnormal hemoglobin** protein or the described disease phenotype.
*Splice site*
- A splice site mutation occurs at the **splice junctions** of introns and exons, leading to errors in mRNA processing.
- This can result in **incorrect protein synthesis** due to exon skipping or intron retention, but it typically does not cause the specific amino acid substitution seen in sickle cell disease.
*Nonsense mutation*
- A nonsense mutation is a point mutation that results in a **premature stop codon**, leading to a **truncated, non-functional protein**.
- While this can cause severe disease, it would typically lead to a complete absence or severe deficiency of functional hemoglobin rather than a structurally altered hemoglobin like HbS.
*Frameshift mutation*
- A frameshift mutation involves the **insertion or deletion of nucleotides** (not in multiples of three), which shifts the reading frame of the mRNA.
- This typically leads to a completely **altered amino acid sequence** downstream of the mutation and usually results in a premature stop codon, leading to a non-functional protein rather than a specific single amino acid substitution.
Promoters and transcription factors US Medical PG Question 5: A mutant stem cell was created by using an inducible RNAi system, such that when doxycycline is added, the siRNA targeting DNA helicase is expressed, effectively knocking down the gene for DNA helicase. Which of the following will occur during DNA replication?
- A. The RNA primer is not created
- B. DNA is not unwound (Correct Answer)
- C. The two melted DNA strands reanneal
- D. DNA supercoiling is not relieved
- E. Newly synthesized DNA fragments are not ligated
Promoters and transcription factors Explanation: ***DNA is not unwound***
- **DNA helicase** is essential for unwinding the **double-stranded DNA** helix, separating it into two single strands. This process creates the **replication fork**.
- Without functional DNA helicase due to **gene knockdown**, the DNA helix cannot be unwound, thus halting DNA replication.
*The RNA primer is not created*
- **RNA primers** are synthesized by **primase**, an enzyme distinct from DNA helicase.
- While unwinding is necessary for primer synthesis, the *creation* of the primer itself is a function of primase.
*The two melted DNA strands reanneal*
- **Reannealing** of DNA strands is prevented by **single-strand binding proteins (SSBs)**, which bind to the separated single strands.
- While helicase unwinds, SSBs specifically keep the strands apart to allow DNA polymerase access.
*DNA supercoiling is not relieved*
- **DNA supercoiling** is relieved by **topoisomerases**, enzymes that cut, unwind, and religate DNA strands to reduce torsional stress.
- This is a distinct function from DNA helicase, which focuses on breaking hydrogen bonds between strands.
*Newly synthesized DNA fragments are not ligated*
- **Ligation** of newly synthesized **Okazaki fragments** on the lagging strand is performed by **DNA ligase**.
- This process occurs downstream from the unwinding step facilitated by DNA helicase.
Promoters and transcription factors US Medical PG Question 6: An investigator is studying the rate of multiplication of hepatitis C virus in hepatocytes. The viral genomic material is isolated, enzymatically cleaved into smaller fragments and then separated on a formaldehyde agarose gel membrane. Targeted probes are then applied to the gel and visualized under x-ray. Which of the following is the most likely structure being identified by this test?
- A. Lipid-linked oligosaccharides
- B. Transcription factors
- C. Polypeptides
- D. Ribonucleic acids (Correct Answer)
- E. Deoxyribonucleic acids
Promoters and transcription factors Explanation: ***Ribonucleic acids***
- The description of isolating "viral genomic material," which is then "enzymatically cleaved" and run on a "formaldehyde agarose gel," followed by the application of "targeted probes" and X-ray visualization, perfectly matches the technique of **Northern blotting**.
- Northern blotting is used to detect and quantify specific **RNA sequences**, which is consistent with the hepatitis C virus being an RNA virus.
*Lipid-linked oligosaccharides*
- These molecules are involved in protein glycosylation and are typically analyzed using techniques like **mass spectrometry** or **chromatography**, not Northern blotting.
- They are not nucleic acid material, which is implied by "viral genomic material" and enzymatic cleavage steps.
*Transcription factors*
- **Transcription factors** are proteins that regulate gene expression and would typically be identified using techniques like **Western blotting** (for protein detection) or Electrophoretic Mobility Shift Assay (EMSA) for DNA binding.
- They are not directly "genomic material" that would be cleaved and run on an agarose gel in this manner.
*Polypeptides*
- **Polypeptides** are chains of amino acids, i.e., proteins, which are normally detected using **Western blotting** after separation on an SDS-PAGE gel.
- The use of "formaldehyde agarose gel" and "enzymatic cleavage" points specifically to nucleic acid analysis, not protein analysis.
*Deoxyribonucleic acids*
- While DNA is genomic material and is often analyzed similarly, the use of a **formaldehyde agarose gel** is characteristic of RNA electrophoresis because formaldehyde prevents RNA from forming secondary structures.
- Furthermore, hepatitis C is a **single-stranded RNA virus**, meaning its genome is RNA, not DNA.
Promoters and transcription factors US Medical PG Question 7: A 24-year-old male presents to the emergency room with a cough and shortness of breath for the past 3 weeks. You diagnose Pneumocystis jiroveci pneumonia (PCP). An assay of the patient's serum reveals the presence of viral protein p24. Which of the following viral genes codes for this protein?
- A. gag (Correct Answer)
- B. pol
- C. rev
- D. env
- E. tat
Promoters and transcription factors Explanation: ***gag***
- The **gag gene** (group-specific antigen) in HIV codes for structural proteins of the virus, including **p24**, which forms the viral capsid.
- The presence of **p24 protein** in the serum is a key marker for **HIV infection**, particularly in the early stages, as it indicates active viral replication.
*pol*
- The **pol gene** codes for essential viral enzymes such as **reverse transcriptase**, **integrase**, and **protease**, which are crucial for the HIV life cycle.
- While vital for viral replication, the **pol gene products** are enzymes involved in processing and replication, not the structural capsid protein p24.
*rev*
- The **rev gene** (regulator of expression of virion proteins) codes for the **Rev protein**, which regulates the export of HIV mRNAs from the nucleus to the cytoplasm.
- This regulatory protein ensures the efficient synthesis of structural and enzymatic proteins but does not directly code for the p24 capsid protein.
*env*
- The **env gene** (envelope) codes for the viral envelope glycoproteins **gp160**, which is cleaved into **gp120** and **gp41**.
- These proteins are critical for viral entry into host cells by binding to CD4 receptors and co-receptors, but they are distinct from the p24 capsid protein.
*tat*
- The **tat gene** (trans-activator of transcription) codes for the **Tat protein**, a powerful trans-activator that enhances the transcription of HIV RNA.
- Tat plays a crucial role in increasing the efficiency of viral gene expression but does not code for structural components like the p24 capsid.
Promoters and transcription factors US Medical PG Question 8: The lac operon allows E. coli to effectively utilize lactose when it is available, and not to produce unnecessary proteins. Which of the following genes is constitutively expressed and results in the repression of the lac operon?
- A. LacY
- B. LacI (Correct Answer)
- C. LacZ
- D. CAP
- E. LacA
Promoters and transcription factors Explanation: ***LacI***
- The **LacI gene** encodes the **Lac repressor protein**, which is constitutively expressed (always produced) and binds to the operator region of the lac operon.
- When bound, the **Lac repressor** blocks RNA polymerase from transcribing the structural genes (LacZ, LacY, LacA), thereby repressing the operon in the absence of lactose.
*LacY*
- The **LacY gene** encodes **lactose permease**, an enzyme responsible for transporting lactose into the bacterial cell.
- Its expression is regulated by the lac operon and is not constitutively expressed; rather, it is induced in the presence of lactose.
*LacZ*
- The **LacZ gene** encodes **beta-galactosidase**, the enzyme that breaks down lactose into glucose and galactose.
- Like LacY, its expression is part of the lac operon and is induced when lactose is available, not expressed constitutively.
*CAP*
- **CAP (Catabolite Activator Protein)** is a regulatory protein that, when bound to cAMP, activates transcription of the lac operon when glucose is absent.
- While essential for lac operon regulation, CAP is not a gene whose constitutive expression leads to repression of the operon.
*LacA*
- The **LacA gene** encodes **thiogalactoside transacetylase**, an enzyme with a less clear role in lactose metabolism but is part of the lac operon.
- Its expression is also regulated and induced along with LacZ and LacY, not constitutively expressed to repress the operon.
Promoters and transcription factors US Medical PG Question 9: A 13-year-old boy is brought to his pediatrician for evaluation of leg pain. Specifically, he has been having pain around his right knee that has gotten progressively worse over the last several months. On presentation, he has swelling and tenderness over his right distal femur. Radiographs are obtained and the results are shown in figure A. His family history is significant in that several family members also had this disorder and others had pathology in the eye near birth. The patient is referred for a genetic consult, and a mutation is found on a certain chromosome. The chromosome that is most likely affected also contains a gene that is associated with which of the following pathologies?
- A. Pancreatic cancers
- B. Colorectal cancer
- C. Soft tissue sarcomas
- D. Neurofibromas
- E. Breast cancer (Correct Answer)
Promoters and transcription factors Explanation: ***Breast cancer***
- This clinical scenario describes hereditary retinoblastoma, characterized by **osteosarcoma** (leg pain, distal femur swelling, and tenderness in a young boy), **retinoblastoma** (eye pathology near birth), and a family history.
- The gene mutated in hereditary retinoblastoma is the **RB1 gene**, located on **chromosome 13**. This chromosome also contains the **BRCA2 gene**, which is associated with an increased risk of **breast cancer**.
*Pancreatic cancers*
- While certain genetic mutations can increase the risk of pancreatic cancer (e.g., **BRCA1/2**, PALB2, ATM), the specific gene and chromosome linked to the described primary condition (hereditary retinoblastoma/osteosarcoma) do not directly point to pancreatic cancer as the most strongly associated co-pathology on that same chromosome in the context of the given options.
- The RB1 gene is on chromosome 13, and while BRCA2 (also on chromosome 13) can be associated with pancreatic cancer, breast cancer is a more prominent association for BRCA2.
*Colorectal cancer*
- Colorectal cancer is often associated with mutations in genes like **APC** (familial adenomatous polyposis) on chromosome 5 or **MLH1**, **MSH2** (Lynch syndrome) on other chromosomes, not primarily chromosome 13 in the context of retinoblastoma.
- There is no direct strong link between the RB1 gene/chromosome 13 and colorectal cancer that would make it the most likely answer among the choices.
*Soft tissue sarcomas*
- While retinoblastoma patients have an increased risk of subsequent cancers, including soft tissue sarcomas, the question asks about a pathology associated with a gene on the **same chromosome** as RB1.
- No major gene causing soft tissue sarcomas is primarily located on chromosome 13 and as prominently linked as BRCA2 to breast cancer.
*Neurofibromas*
- Neurofibromas are characteristic of **neurofibromatosis type 1 (NF1)**, caused by a mutation in the **NF1 gene** located on chromosome 17.
- This is a different chromosome and gene entirely from the RB1 gene on chromosome 13.
Promoters and transcription factors US Medical PG Question 10: An investigator is studying the effects of zinc deprivation on cancer cell proliferation. It is hypothesized that because zinc is known to be a component of transcription factor motifs, zinc deprivation will result in slower tumor growth. To test this hypothesis, tumor cells are cultured on media containing low and high concentrations of zinc. During the experiment, a labeled oligonucleotide probe is used to identify the presence of a known transcription factor. The investigator most likely used which of the following laboratory techniques?
- A. ELISA
- B. PCR
- C. Western blot
- D. Northern blot
- E. Southwestern blot (Correct Answer)
Promoters and transcription factors Explanation: ***Southwestern blot***
- A **Southwestern blot** specifically identifies **DNA-binding proteins** (such as transcription factors) by detecting their ability to bind to specific **labeled DNA oligonucleotide probes**
- The technique involves: protein separation by gel electrophoresis → transfer to membrane → probing with **labeled double-stranded DNA oligonucleotide**
- This directly answers the question: using a labeled oligonucleotide probe to identify a transcription factor
*ELISA*
- **ELISA** detects and quantifies proteins using **antibody-antigen interactions**, not DNA-binding activity
- While it could detect the presence of a transcription factor protein, it cannot assess the protein's ability to bind to specific DNA sequences
- Does not utilize oligonucleotide probes for detection
*PCR*
- **PCR** amplifies specific **DNA sequences** but does not detect or characterize proteins
- This technique would amplify DNA, not identify DNA-binding proteins
- Not applicable for detecting transcription factor presence or function
*Western blot*
- **Western blot** detects specific proteins using **antibodies**, not oligonucleotide probes
- While it could confirm transcription factor protein presence, it cannot assess DNA-binding capability
- Uses antibody-based detection, not nucleotide probe-based detection
*Northern blot*
- **Northern blot** detects specific **RNA molecules**, not DNA-binding proteins
- Uses labeled DNA or RNA probes to detect RNA, not to detect proteins that bind DNA
- Wrong target molecule (RNA vs. proteins)
More Promoters and transcription factors US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.