Post-transcriptional modifications US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Post-transcriptional modifications. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Post-transcriptional modifications US Medical PG Question 1: An investigator is developing a drug for muscle spasms. The drug inactivates muscular contraction by blocking the site where calcium ions bind to regulate actin-myosin interaction. Which of the following is the most likely site of action of this drug?
- A. Troponin C (Correct Answer)
- B. Myosin-binding site
- C. Acetylcholine receptor
- D. Ryanodine receptor
- E. Myosin head
Post-transcriptional modifications Explanation: ***Troponin C***
- **Calcium ions** bind to **Troponin C**, initiating a conformational change in the troponin-tropomyosin complex, which exposes the **myosin-binding sites on actin**.
- Blocking this site directly prevents the **calcium-mediated regulation** of muscle contraction, thus inactivating it.
*Myosin-binding site*
- The **myosin-binding site** is located on the **actin filament** and is where the **myosin head** attaches to form cross-bridges.
- While essential for contraction, this site doesn't directly bind calcium ions to initiate the process.
*Acetylcholine receptor*
- The **acetylcholine receptor** is located on the **neuromuscular junction** and mediates the transmission of a nerve impulse to the muscle fiber.
- Blocking this receptor would prevent muscle depolarization, but it's not the direct site where calcium ions regulate actin-myosin interaction.
*Ryanodine receptor*
- The **ryanodine receptor** is located on the **sarcoplasmic reticulum** and controls the release of calcium ions into the sarcoplasm.
- While it's involved in calcium signaling, it doesn't represent the site where calcium binds to *regulate* the actin-myosin interaction itself.
*Myosin head*
- The **myosin head** contains the **ATPase activity** and binds to actin to form cross-bridges, enabling muscle contraction.
- It does not directly bind **calcium ions** to regulate the initiation of contraction; instead, its binding to actin is regulated by the troponin-tropomyosin complex.
Post-transcriptional modifications US Medical PG Question 2: A 25-year-old female comes to the clinic complaining of fatigue and palpitations. She has been undergoing immense stress from her thesis defense and has been extremely tired. The patient denies any weight loss, diarrhea, cold/heat intolerance. TSH was within normal limits. She reports a family history of "blood disease" and was later confirmed positive for β-thalassemia minor. It is believed that abnormal splicing of the beta globin gene results in β-thalassemia. What is removed during this process that allows RNA to be significantly shorter than DNA?
- A. 3'-poly(A) tail
- B. Exons
- C. Introns (Correct Answer)
- D. microRNAs
- E. snRNPs
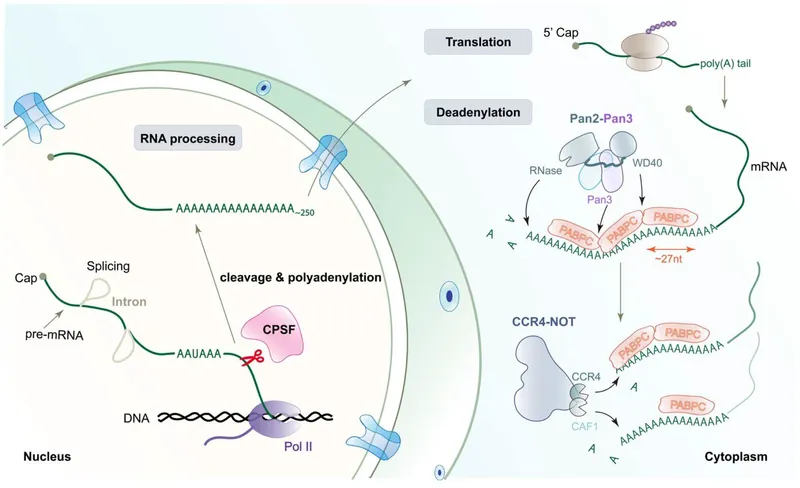
Post-transcriptional modifications Explanation: **Introns**
- **Introns** are non-coding regions within a gene that are removed from the pre-mRNA transcript during **splicing**.
- This removal and the subsequent ligation of exons lead to a mature mRNA molecule that is significantly shorter than the initial DNA template.
*3'-poly(A) tail*
- The **3'-poly(A) tail** is an addition to the 3' end of the mRNA molecule, not a removed segment during splicing, and it provides stability and aids in translation.
- While it contributes to mRNA processing, its addition does not involve removing existing sequences to shorten the transcript.
*Exons*
- **Exons** are the coding regions of a gene that are retained and ligated together to form the mature mRNA, which is then translated into protein.
- If exons were removed, the resulting protein would be truncated or non-functional, and the mRNA would not contain the necessary genetic information.
*microRNAs*
- **MicroRNAs (miRNAs)** are small non-coding RNA molecules that regulate gene expression by inhibiting translation or promoting mRNA degradation.
- They are not part of the pre-mRNA transcript that is processed into mRNA; rather, they are distinct regulatory molecules.
*snRNPs*
- **Small nuclear ribonucleoproteins (snRNPs)** are components of the spliceosome, the molecular machine responsible for carrying out splicing.
- They are involved in the process of intron removal but are not themselves removed from the RNA; they are catalytic machinery.
Post-transcriptional modifications US Medical PG Question 3: An investigator studying protein synthesis in human stem cells isolates tRNA molecules bound to mRNA molecules. The isolated tRNA molecules have inosine in the 5' position of the anticodon; of these, some are bound to adenine, some to cytosine, and some to uracil at the 3' position of the mRNA codon. Which of the following properties of the genetic code is best illustrated by this finding?
- A. Unambiguity
- B. Non-overlapping
- C. Degeneracy (Correct Answer)
- D. Specificity of the start codon
- E. Specificity of stop codons
Post-transcriptional modifications Explanation: ***Degeneracy***
- The finding that a single tRNA anticodon (with **inosine** at the 5' position) can bind to multiple different mRNA codons (ending in **adenine, cytosine, or uracil**) illustrates the concept of **degeneracy** in the genetic code.
- This **wobble hypothesis** allows fewer tRNAs to recognize more than one codon for a given amino acid, meaning multiple codons can code for the same amino acid.
*Unambiguity*
- The genetic code is unambiguous, meaning that each codon specifies **only one specific amino acid** (or a stop signal) and never two different amino acids.
- This finding, however, shows one tRNA recognizing multiple codons, not one codon coding for multiple amino acids.
*Non-overlapping*
- The **non-overlapping** nature of the genetic code means that each nucleotide in an mRNA sequence is read only once as part of a single codon, without sharing nucleotides between adjacent codons.
- This concept describes how codons are read sequentially, not the flexibility of codon-anticodon pairing.
*Specificity of the start codon*
- The **start codon (AUG)** specifically initiates translation, coding for methionine, and signals the beginning of a polypeptide chain.
- This finding relates to the wobble pairing at the 3' end of the codon, not the initiation of translation.
*Specificity of stop codons*
- **Stop codons (UAA, UAG, UGA)** specifically signal the termination of translation without coding for any amino acid.
- This finding describes the flexibility of codon-anticodon pairing, not the distinct function of termination codons.
Post-transcriptional modifications US Medical PG Question 4: A 25-year-old woman presents into the clinic complaining of worsening malaise, hair loss, and a rash on her face. The patient states that she has been avoiding daylight because the rash becomes painful, and she has not been able to go to classes because of debilitating arthralgia in her fingers and ankles. No significant past medical history. She takes no medication. At the time of the consult, the patient has a fever of 39.0°C (102.2 °F). The presence of which of the following is most commonly seen on diagnostic labs in this patient’s most likely condition?
- A. Anti-smith antibody
- B. Anti-dsDNA
- C. Anti-Ro antibody
- D. Antinuclear antibody (Correct Answer)
- E. Anti-histone antibody
Post-transcriptional modifications Explanation: ***Antinuclear antibody***
- The patient's symptoms (malaise, hair loss, photosensitive malar rash, arthralgia, fever) are highly suggestive of **Systemic Lupus Erythematosus (SLE)**.
- An **antinuclear antibody (ANA)** test is the **most sensitive screening test** for SLE and is present in over 95% of patients with the condition.
*Anti-dsDNA*
- While **anti-dsDNA antibodies** are highly specific for SLE and often correlate with disease activity, particularly **lupus nephritis**, they are present in only about 70-80% of SLE patients, making them less common than ANA.
- This antibody is more specific than ANA but **not as sensitive** as ANA for initial diagnosis.
*Anti-smith antibody*
- **Anti-Smith antibodies** are highly specific for SLE (pathognomonic), meaning their presence strongly indicates SLE, but they are found in only about 20-30% of SLE patients, making them relatively uncommon.
- Their presence is **not as common** as ANA or even anti-dsDNA in the general SLE population.
*Anti-Ro antibody*
- **Anti-Ro/SSA antibodies** are associated with specific manifestations of SLE, such as **subacute cutaneous lupus erythematosus**, neonatal lupus, and Sjögren's syndrome, but are not present in all SLE patients.
- This antibody is present in approximately 25-30% of SLE patients, and its diagnostic significance is more often for specific subsets rather than overall disease presence.
*Anti-histone antibody*
- **Anti-histone antibodies** are primarily associated with **drug-induced lupus (DIL)**, although they can also be present in a small percentage of idiopathic SLE cases.
- Given that the patient takes no medication and her symptoms are more consistent with idiopathic SLE, this antibody is **less likely to be the most commonly seen** diagnostic marker.
Post-transcriptional modifications US Medical PG Question 5: An investigator is studying the crossbridge cycle of muscle contraction. Tissue from the biceps brachii muscle is obtained at the autopsy of an 87-year-old man. Investigation of the muscle tissue shows myosin heads attached to actin filaments. Binding of myosin heads to which of the following elements would most likely cause detachment of myosin from actin filaments?
- A. ATP (Correct Answer)
- B. Troponin C
- C. Tropomyosin
- D. ADP
- E. cGMP
Post-transcriptional modifications Explanation: ***ATP***
- The binding of **ATP** to the **myosin head** causes a conformational change that reduces its affinity for actin, leading to detachment.
- This step is crucial for the muscle to relax and for the subsequent power stroke to occur.
*Troponin C*
- **Troponin C** is a regulatory protein that binds calcium, which then causes a conformational change in the troponin-tropomyosin complex, revealing the **actin binding sites** for myosin.
- It does not directly cause myosin detachment; instead, it facilitates the binding of myosin to actin.
*Tropomyosin*
- **Tropomyosin** is a long, fibrous protein that covers the **myosin-binding sites** on actin in a relaxed muscle, preventing cross-bridge formation.
- Its movement, regulated by troponin, allows myosin to bind, but it does not directly cause detachment.
*ADP*
- **ADP** is released from the myosin head during the power stroke, but its binding does not cause detachment; rather, it is present during the strongly bound state before **ATP** binds.
- The presence of **ADP** and inorganic phosphate (Pi) often promotes the strong binding of myosin to actin.
*cGMP*
- **cGMP** (cyclic guanosine monophosphate) is a second messenger involved in various cellular processes, including smooth muscle relaxation, but it is not directly involved in the cross-bridge cycle and detachment of **myosin from actin** in skeletal muscle.
- Its primary role in muscle physiology is often linked to nitric oxide signaling and vasodilation.
Post-transcriptional modifications US Medical PG Question 6: A 30-year-old African American woman develops a facial rash in a "butterfly" pattern over her face and complains of feeling tired and achy in her joints. In the course of a full rheumatologic workup you note that she has anti-snRNP antibodies. Which of the following do snRNPs affect?
- A. Transcription of mRNA
- B. Intron removal from the mRNA (Correct Answer)
- C. Protection of mRNA from degradation
- D. Polyadenylation of the 3' end of mRNA
- E. Addition of the 5' 7-methylguanosine cap of mRNA
Post-transcriptional modifications Explanation: ***Intron removal from the mRNA***
- **Small nuclear ribonucleoproteins (snRNPs)** are crucial components of the **spliceosome**, the molecular machinery responsible for removing non-coding introns from pre-mRNA.
- **snRNPs** recognize and bind to specific sequences within introns and at exon-intron junctions, guiding the splicing process to produce mature mRNA.
*Transcription of mRNA*
- **Transcription** is the process where DNA is copied into RNA, primarily catalyzed by **RNA polymerase**.
- While snRNPs are involved in post-transcriptional modification, they do not directly affect the initial synthesis of the mRNA transcript.
*Protection of mRNA from degradation*
- The **poly-A tail** and the **5' cap** play significant roles in protecting mRNA from degradation by exonucleases.
- While splicing is essential for producing a functional message, snRNPs themselves are not primarily involved in the degradation protection mechanism.
*Polyadenylation of the 3' end of mRNA*
- **Polyadenylation** involves the addition of a **poly-A tail** to the 3' end of the mRNA, which is mediated by poly-A polymerase.
- This process is distinct from splicing and occurs after the mature mRNA has been formed.
*Addition of the 5' 7-methylguanosine cap of mRNA*
- The **5' cap**, a 7-methylguanosine residue, is added to the 5' end of the mRNA during transcription and is crucial for ribosome binding and mRNA stability.
- This capping process occurs early in mRNA synthesis and is not directly mediated by snRNPs.
Post-transcriptional modifications US Medical PG Question 7: An investigator is studying the genotypes of wingless fruit flies using full exome sequencing. Compared to wild-type winged fruit flies, the wingless fruit flies are found to have a point mutation in the gene encoding wing bud formation during embryogenesis. The point mutation in the gene causes the mRNA transcript to have a 'UUG' segment instead of an 'AUG' segment. Which of the following processes is most likely affected by this mutation?
- A. Cleavage of 5' intron
- B. Binding of met-tRNA to 40S complex (Correct Answer)
- C. Catalyzation of peptide bond formation
- D. Dissociation of mRNA from ribosome complex
- E. Shift of peptidyl-tRNA from A to P site
Post-transcriptional modifications Explanation: ***Binding of met-tRNA to 40S complex***
- The **start codon AUG** is essential for the initiation of translation, as it signals where the ribosome should begin synthesizing the polypeptide chain and recruits the initiator tRNA carrying **methionine (met-tRNA)** to the 40S ribosomal subunit.
- A mutation from **AUG to UUG** means the ribosome will not recognize the correct start site, preventing the initial binding of met-tRNA and the formation of the **initiation complex**.
*Cleavage of 5' intron*
- This process is part of **RNA splicing**, which occurs after transcription in the nucleus, where introns are removed from the **pre-mRNA**.
- The described mutation affects a **codon sequence** in the mRNA, which is a post-splicing event related to translation, not intron cleavage.
*Catalyzation of peptide bond formation*
- This occurs during the **elongation phase of translation**, where the peptidyl transferase activity of the ribosome forms peptide bonds between amino acids.
- The mutation prevents the **initiation of translation** altogether, meaning elongation and peptide bond formation will not even begin.
*Dissociation of mRNA from ribosome complex*
- This event happens at the **termination phase of translation**, when a stop codon is reached, and release factors cause the ribosome to dissociate from the mRNA and the newly synthesized polypeptide.
- The mutation prevents the **start of translation**, so the ribosome will not reach the stage where it would dissociate from the mRNA.
*Shift of peptidyl-tRNA from A to P site*
- This is a step in the **elongation phase of translation**, specifically the **translocation process**, where the ribosome moves along the mRNA, shifting the peptidyl-tRNA from the A (aminoacyl) site to the P (peptidyl) site.
- Since the **initiation of translation** is blocked by the mutated start codon, the ribosome cannot begin polypeptide synthesis, and thus, elongation steps like translocation cannot occur.
Post-transcriptional modifications US Medical PG Question 8: An investigator is studying the function of the endoplasmic reticulum in genetically modified lymphocytes. A gene is removed that facilitates the binding of ribosomes to the endoplasmic reticulum. Which of the following processes is most likely to be impaired as a result of this genetic modification?
- A. Production of secretory proteins (Correct Answer)
- B. Neutralization of toxins
- C. Ubiquitination of proteins
- D. α-Oxidation of fatty acids
- E. Synthesis of ketone bodies
Post-transcriptional modifications Explanation: ***Production of secretory proteins***
- Ribosomes bound to the **rough endoplasmic reticulum (RER)** are responsible for synthesizing proteins destined for secretion, insertion into membranes, or delivery to organelles like lysosomes.
- If ribosomes cannot bind to the ER, these proteins will be synthesized in the **cytosol** and lack the proper signals and processing for their intended destination and function.
*Neutralization of toxins*
- The **smooth endoplasmic reticulum (SER)**, not the RER, is primarily involved in **detoxification** processes, particularly drug metabolism and neutralization of toxins.
- This function relies on enzymes embedded within the SER membrane and is largely independent of ribosome binding.
*Ubiquitination of proteins*
- **Ubiquitination** is a post-translational modification that tags proteins for degradation by the **proteasome** or for trafficking to specific cellular compartments.
- This process occurs primarily in the **cytosol** and does not directly rely on ribosome binding to the ER for protein synthesis.
*α-Oxidation of fatty acids*
- **α-oxidation of fatty acids** is a metabolic pathway that occurs primarily in the **peroxisomes**.
- It is distinct from protein synthesis on the ER and would not be directly impacted by the inability of ribosomes to bind to the ER.
*Synthesis of ketone bodies*
- The **synthesis of ketone bodies** (ketogenesis) primarily occurs in the **mitochondria** of liver cells.
- This metabolic pathway is not directly dependent on ribosome binding to the endoplasmic reticulum for its function.
Post-transcriptional modifications US Medical PG Question 9: E. coli has the ability to regulate its enzymes to break down various sources of energy when available. It prevents waste by the use of the lac operon, which encodes a polycistronic transcript. At a low concentration of glucose and absence of lactose, which of the following occurs?
- A. Decreased cAMP levels result in poor binding to the catabolite activator protein
- B. Increased cAMP levels result in binding to the catabolite activator protein (Correct Answer)
- C. Increased allolactose levels bind to the repressor
- D. Repressor releases from lac operator
- E. Transcription of the lac Z, Y, and A genes increase
Post-transcriptional modifications Explanation: ***Increased cAMP levels result in binding to the catabolite activator protein***
- In the absence of glucose, **adenylate cyclase** activity increases, leading to higher levels of **cAMP**.
- **cAMP** then binds to the **catabolite activator protein (CAP)**, forming the **cAMP-CAP complex**, which is crucial for activating lac operon transcription in the absence of glucose.
*Decreased cAMP levels result in poor binding to the catabolite activator protein*
- **Decreased glucose levels** actually lead to **increased cAMP** synthesis, not decreased.
- High **cAMP** levels enhance, not hinder, its binding to **CAP**.
*Increased allolactose levels bind to the repressor*
- **Allolactose** is an inducer that forms in the presence of **lactose**, which is stated to be absent in this scenario.
- Therefore, **allolactose levels** would be low, and it would not bind to the **repressor**.
*Repressor releases from lac operator*
- The **repressor protein** is bound to the **lac operator** in the absence of lactose.
- For the **repressor to be released**, **allolactose** (formed from lactose) must be present to bind to it.
*Transcription of the lac Z, Y, and A genes increase*
- While **cAMP-CAP binding** would promote transcription, the **absence of lactose** means the **repressor remains bound** to the operator.
- This binding effectively blocks RNA polymerase, preventing significant transcription of the **lac Z, Y, and A genes**, regardless of high **cAMP** levels.
Post-transcriptional modifications US Medical PG Question 10: Expression of an mRNA encoding for a soluble form of the Fas protein prevents a cell from undergoing programmed cell death. However, after inclusion of a certain exon, this same Fas pre-mRNA eventually leads to the translation of a protein that is membrane bound, subsequently promoting the cell to undergo apoptosis. Which of the following best explains this finding?
- A. Histone deacetylation
- B. DNA missense mutation
- C. Alternative splicing (Correct Answer)
- D. Base excision repair
- E. Post-translational modifications
Post-transcriptional modifications Explanation: ***Alternative splicing***
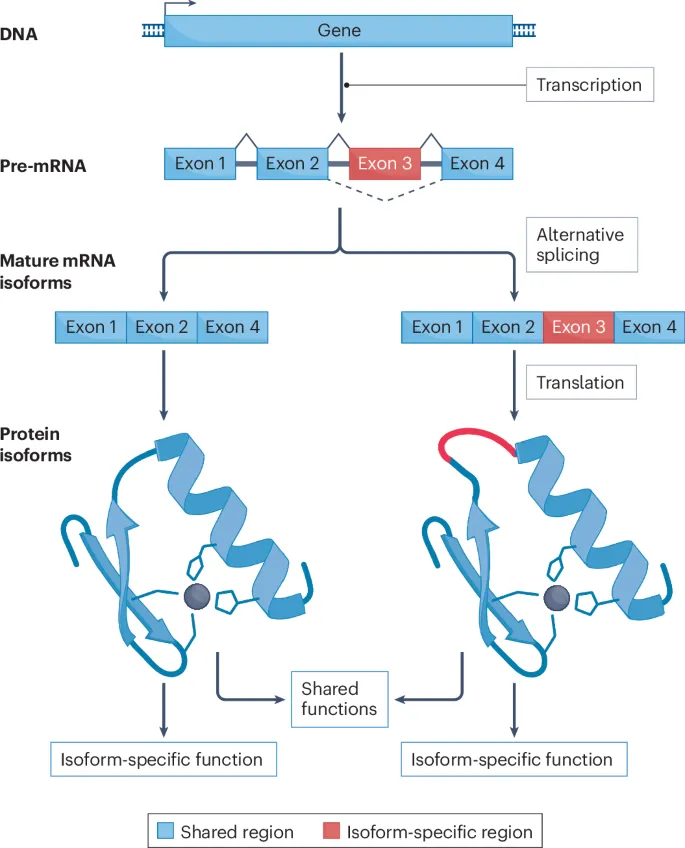
- The scenario describes a single **pre-mRNA** producing two different protein forms (soluble vs. membrane-bound Fas) with distinct functions, depending on the inclusion or exclusion of a specific **exon**. This is the hallmark of alternative splicing.
- **Alternative splicing** allows a single gene to encode multiple protein isoforms, leading to diverse cellular functions and regulation.
*Histone deacetylation*
- **Histone deacetylation** is a mechanism of gene regulation that typically represses gene expression by making DNA less accessible for transcription, not by altering the protein product of an already transcribed gene.
- It affects whether a gene is turned "on" or "off," but doesn't explain how the same pre-mRNA produces different protein versions.
*DNA missense mutation*
- A **DNA missense mutation** would alter a single base pair in the DNA, potentially changing one amino acid in the resulting protein.
- While it can lead to functional changes in a protein, it would not explain the complete inclusion or exclusion of an entire exon, which profoundly changes the protein's overall structure and membrane association in this manner.
*Base excision repair*
- **Base excision repair** is a DNA repair pathway that corrects small, non-bulky DNA lesions, such as damaged or modified bases.
- This process is involved in maintaining genomic integrity and does not explain the differential processing of an mRNA transcript to produce two distinct protein isoforms.
*Post-translational modifications*
- **Post-translational modifications** (PTMs) occur after protein translation and involve chemical changes to the protein (e.g., phosphorylation, glycosylation).
- While PTMs can alter protein function or localization, they do not explain how an entire exon's inclusion or exclusion leads to fundamentally different protein structures (soluble vs. membrane-bound).
More Post-transcriptional modifications US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.