Transcription/translation

On this page
🧬 The Molecular Assembly Line: From DNA Blueprint to Protein Product
Every protein in your body-from the hemoglobin carrying oxygen through your bloodstream to the antibodies defending against infection-begins as a coded message locked in DNA. You'll trace the complete journey from gene to functional protein, mastering how RNA polymerase reads DNA blueprints during transcription, how ribosomes decode mRNA into amino acid chains during translation, and how nascent proteins fold and modify themselves into their final working forms. Understanding this molecular assembly line reveals not only how cells build themselves but also how mutations derail the process and how clinicians can intervene when transcription or translation goes wrong.
The central dogma of molecular biology-DNA → RNA → Protein-represents one of medicine's most fundamental processes. Every therapeutic intervention, from antibiotics targeting bacterial ribosomes to cancer drugs disrupting transcription, depends on understanding this molecular choreography. Master these pathways, and you unlock the logic behind genetic diseases, antimicrobial mechanisms, and targeted therapies.
📌 Remember: TRANSCRIPTION = DNA → RNA (nucleus), TRANSLATION = RNA → Protein (ribosome)
The transcription-translation axis operates with remarkable efficiency: human cells transcribe approximately 20,000 genes daily, producing 10 million proteins per cell through coordinated molecular machinery. Understanding this process reveals why 85% of genetic diseases result from transcriptional errors, and why 60% of antibiotics target translation machinery.
⭐ Clinical Pearl: Rifampin blocks bacterial RNA polymerase (IC50 = 0.02 μM), while eukaryotic RNA polymerase II remains unaffected (IC50 > 100 μM), explaining its selective antimicrobial action.
| Process Component | Location | Key Enzyme | Clinical Target | Selectivity Index |
|---|---|---|---|---|
| Prokaryotic Transcription | Cytoplasm | RNA Polymerase | Rifampin | 1:5000 |
| Eukaryotic Transcription | Nucleus | RNA Pol II | α-Amanitin | 1:1 |
| Prokaryotic Translation | Cytoplasm | 70S Ribosome | Streptomycin | 1:500 |
| Eukaryotic Translation | Cytoplasm/ER | 80S Ribosome | Cycloheximide | 1:1 |
| Mitochondrial Translation | Mitochondria | 70S-like | Chloramphenicol | 1:10 |
Connect this foundational understanding through the transcriptional machinery architecture to understand how molecular precision enables therapeutic selectivity.
🧬 The Molecular Assembly Line: From DNA Blueprint to Protein Product
⚙️ The Transcriptional Engine: RNA Polymerase Command Center
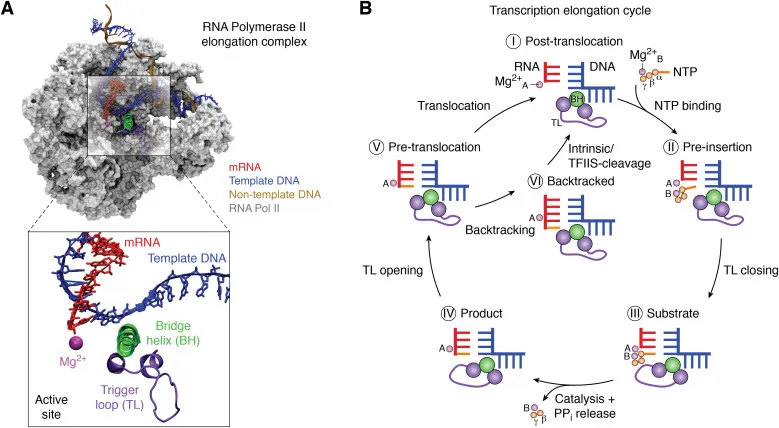
RNA polymerase functions as a molecular motor with distinct operational phases: initiation, elongation, and termination. The enzyme's active site accommodates double-stranded DNA through a 27-Å channel, while the bridge helix coordinates nucleotide addition with translocation mechanics.
📌 Remember: PINE - Promoter recognition, Initiation complex, Nucleotide addition, Elongation cycle
Prokaryotic RNA polymerase contains 5 core subunits (α₂ββ'ω) plus sigma factors for promoter recognition, while eukaryotic systems employ 3 specialized polymerases: Pol I (rRNA), Pol II (mRNA), and Pol III (tRNA). This specialization enables tissue-specific regulation and developmental control.
⭐ Clinical Pearl: α-Amanitin (death cap mushroom toxin) specifically inhibits RNA Pol II at nanomolar concentrations, causing hepatic necrosis within 48-72 hours through mRNA synthesis blockade.
| RNA Polymerase | Target Genes | Promoter Elements | Inhibitor Sensitivity | Clinical Relevance |
|---|---|---|---|---|
| Pol I | rRNA (18S, 28S) | UBF, SL1 | α-Amanitin resistant | Nucleolar stress |
| Pol II | mRNA, miRNA | TFIID, TFIIB | α-Amanitin sensitive | Drug target |
| Pol III | tRNA, 5S rRNA | TFIIIB, TFIIIC | α-Amanitin resistant | tRNA disorders |
| Bacterial | All genes | Sigma factors | Rifampin sensitive | Antibiotic target |
| Mitochondrial | mtDNA genes | mtTFA | Ethidium bromide | Mitochondrial disease |
Connect this enzymatic precision through the elongation and termination mechanisms to understand how transcriptional control regulates gene expression.
⚙️ The Transcriptional Engine: RNA Polymerase Command Center
🎯 The Elongation Marathon: Transcriptional Processivity Control
📌 Remember: STEP - Stalling sites, Termination signals, Elongation factors, Pausing mechanisms
Prokaryotic elongation operates at 40-50 nucleotides/second with intrinsic termination (hairpin structures) or Rho-dependent termination. Eukaryotic elongation proceeds at 25-30 nucleotides/second with complex regulatory mechanisms including chromatin remodeling and co-transcriptional processing.
- Elongation Control Mechanisms
- Pausing sites: TFIIS-mediated rescue from arrested complexes
- Chromatin barriers: Histone modifications regulate polymerase progression
- H3K36me3: elongation-permissive chromatin state
- H3K27me3: elongation-inhibitory chromatin state
- Co-transcriptional processing: 5' capping occurs after 20-30 nucleotides
- 7-methylguanosine cap addition within seconds of initiation
⭐ Clinical Pearl: Thalassemia mutations often involve premature termination during α- or β-globin transcription, with nonsense-mediated decay eliminating >90% of aberrant transcripts.
| Termination Type | Mechanism | Efficiency | Clinical Example | Therapeutic Target |
|---|---|---|---|---|
| Intrinsic (Prokaryotic) | Hairpin + U-rich | 85-95% | rRNA operons | Antisense oligonucleotides |
| Rho-dependent | Helicase action | 95-99% | Prophage control | Rho inhibitors |
| Polyadenylation | AAUAAA signal | >99% | mRNA 3' processing | Splicing modulators |
| Collision | Replication fork | Variable | DNA damage | Checkpoint inhibitors |
| Chromatin | Heterochromatin | 60-80% | X-inactivation | Epigenetic drugs |
| %%{init: {'flowchart': {'htmlLabels': true}}}%% | ||||
| flowchart TD |
Start["🧬 Elongating Pol
• RNA polymerase II• Active synthesis"]
Signal{"📋 Regulatory Signal
• Checkpoint control• Pathway steering"}
TFIIS["🩺 TFIIS Rescue
• Cleaves backtracked• Restarts synthesis"]
Release["✅ Release Complex
• Dissociation step• mRNA completion"]
RNA["🔬 RNA Processing
• Splicing/Capping• Polyadenylation"]
Remodel["💊 Remodel Req
• Histone modifiers• SWI/SNF complex"]
Barrier{"📋 Barrier Type
• Chromatin state• Access control"}
Resume["👁️ Resume Elongation
• Continue coding• Forward motion"]
Block["⚠️ Transcribe Block
• Arrested state• Gene silenced"]
Start --> Signal
Signal -->|Pause Site| TFIIS Signal -->|Termination| Release Signal -->|Chrom Barrier| Remodel
TFIIS --> Resume Release --> RNA Remodel --> Barrier
Barrier -->|Permissive| Resume Barrier -->|Repressive| Block
style Start fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8 style Signal fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E style TFIIS fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8 style Release fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252 style RNA fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C style Remodel fill:#F1FCF5, stroke:#BEF4D8, stroke-width:1.5px, rx:12, ry:12, color:#166534 style Barrier fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E style Resume fill:#EEFAFF, stroke:#DAF3FF, stroke-width:1.5px, rx:12, ry:12, color:#0369A1 style Block fill:#FDF4F3, stroke:#FCE6E4, stroke-width:1.5px, rx:12, ry:12, color:#B91C1C
> 💡 **Master This**: **Co-transcriptional recruitment** of processing factors ensures **mRNA quality control**-defects in this coupling cause **>40%** of inherited diseases through **splicing errors** or **nonsense-mediated decay**.
Connect this elongation control through the translation initiation machinery to understand how mRNA processing prepares transcripts for protein synthesis.
🎯 The Elongation Marathon: Transcriptional Processivity Control
🔧 The Translation Initiation Complex: Ribosomal Assembly Mastery
📌 Remember: SCAN - Small subunit binding, Cap recognition, AUG identification, Nacent peptide start
The eIF4F complex (eIF4E + eIF4G + eIF4A) recognizes the 5' cap structure, while eIF4B and eIF4H enhance helicase activity for secondary structure resolution. The 43S complex scans 5' → 3' at 4-6 nucleotides/second until AUG recognition.
- Initiation Factor Functions
- eIF2: Met-tRNA delivery in ternary complex with GTP
- eIF2α phosphorylation: stress response blocking 80% of translation
- PERK, PKR, GCN2, HRI kinases respond to different stresses
- eIF4E: Cap-binding protein with nanomolar affinity
- 4E-BP1 phosphorylation releases eIF4E for active translation
- mTOR pathway regulates 4E-BP1 phosphorylation status
- eIF2: Met-tRNA delivery in ternary complex with GTP
⭐ Clinical Pearl: eIF2α phosphorylation during ER stress reduces global protein synthesis by >90% while selectively enhancing ATF4 translation through upstream open reading frames.
| Initiation Factor | Function | Regulation | Disease Association | Drug Target |
|---|---|---|---|---|
| eIF2 | Met-tRNA delivery | Phosphorylation | Vanishing white matter | ISRIB |
| eIF4E | Cap recognition | 4E-BP binding | Cancer overexpression | 4EGI-1 |
| eIF4G | Scaffold protein | Cleavage | Viral shutoff | Protease inhibitors |
| eIF4A | RNA helicase | ATP-dependent | DDX3 mutations | Silvestrol |
| eIF6 | 60S anti-association | Phosphorylation | Shwachman-Diamond | Growth factors |
| %%{init: {'flowchart': {'htmlLabels': true}}}%% | ||||
| flowchart TD |
A["<b>🧬 5' Capped mRNA</b><br><span style='display:block; text-align:left; color:#555'>• Cap-dependent</span><span style='display:block; text-align:left; color:#555'>• mRNA initiation</span>"]
B["<b>🔬 eIF4F Binding</b><br><span style='display:block; text-align:left; color:#555'>• Recognition step</span><span style='display:block; text-align:left; color:#555'>• Complex recruitment</span>"]
C["<b>🧬 43S Preinitiation</b><br><span style='display:block; text-align:left; color:#555'>• Small ribosome unit</span><span style='display:block; text-align:left; color:#555'>• Complex assembly</span>"]
D["<b>🔍 5' UTR Scanning</b><br><span style='display:block; text-align:left; color:#555'>• Processive search</span><span style='display:block; text-align:left; color:#555'>• Find start codon</span>"]
E{"<b>📋 AUG Found?</b><br><span style='display:block; text-align:left; color:#555'>• Start codon check</span><span style='display:block; text-align:left; color:#555'>• Sequence match</span>"}
F["<b>🔁 Continue Scan</b><br><span style='display:block; text-align:left; color:#555'>• Move downstream</span><span style='display:block; text-align:left; color:#555'>• Search continues</span>"]
G["<b>🧬 60S Joining</b><br><span style='display:block; text-align:left; color:#555'>• Large subunit joins</span><span style='display:block; text-align:left; color:#555'>• Factor release</span>"]
H["<b>🧬 80S Ribosome</b><br><span style='display:block; text-align:left; color:#555'>• Complete complex</span><span style='display:block; text-align:left; color:#555'>• Translation ready</span>"]
I["<b>✅ Elongation</b><br><span style='display:block; text-align:left; color:#555'>• Protein synthesis</span><span style='display:block; text-align:left; color:#555'>• Chain extension</span>"]
A --> B
B --> C
C --> D
D --> E
E -->|No| F
F --> D
E -->|Yes| G
G --> H
H --> I
style A fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8
style B fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C
style C fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C
style D fill:#EEFAFF, stroke:#DAF3FF, stroke-width:1.5px, rx:12, ry:12, color:#0369A1
style E fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E
style F fill:#EEFAFF, stroke:#DAF3FF, stroke-width:1.5px, rx:12, ry:12, color:#0369A1
style G fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C
style H fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8
style I fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
> 💡 **Master This**: **Internal ribosome entry sites (IRES)** bypass **cap-dependent initiation** during **cellular stress**, enabling **survival protein synthesis** when **eIF4F** is **inactive**-this mechanism explains **cancer cell survival** during **chemotherapy**.
Connect this initiation precision through the elongation and termination cycles to understand how protein synthesis maintains fidelity and responds to cellular demands.
🔧 The Translation Initiation Complex: Ribosomal Assembly Mastery
⚖️ The Protein Assembly Cycle: Elongation and Termination Precision
📌 Remember: APE sites - Aminoacyl (incoming), Peptidyl (growing chain), Exit (departing tRNA)
eEF1A delivers aminoacyl-tRNA to the A-site with GTP hydrolysis ensuring correct codon-anticodon pairing. eEF2 catalyzes translocation with additional GTP hydrolysis, while peptidyl transferase (23S rRNA in prokaryotes, 28S rRNA in eukaryotes) forms peptide bonds through ribozyme activity.
- Elongation Fidelity Mechanisms
- Initial selection: 100-fold discrimination against incorrect tRNAs
- Induced fit mechanism enhances codon-anticodon accuracy
- GTP hydrolysis provides kinetic proofreading
- Proofreading: Additional 100-fold error reduction
- eEF1A conformational change after GTP hydrolysis
- Incorrect tRNAs dissociate faster than peptide bond formation
- Initial selection: 100-fold discrimination against incorrect tRNAs
⭐ Clinical Pearl: Diphtheria toxin ADP-ribosylates eEF2 at Diphthamide-699, completely blocking protein synthesis and causing cell death within 24-48 hours.
| Elongation Factor | Function | GTP Requirement | Antibiotic Target | Clinical Significance |
|---|---|---|---|---|
| eEF1A | tRNA delivery | 1 GTP/amino acid | Didemnin B | Cancer research |
| eEF2 | Translocation | 1 GTP/amino acid | Diphtheria toxin | Vaccine target |
| eEF1B | GDP/GTP exchange | Catalytic | None known | Essential viability |
| eEF3 | E-site clearance | 1 ATP/cycle | None known | Fungal-specific |
| eIF5A | First peptide bond | Hypusine-dependent | GC7 inhibitor | Cancer target |
💡 Master This: Ribosome recycling requires ABCE1 (ATP-binding cassette protein) to split 80S ribosomes-mutations cause severe growth retardation and developmental delays through ribosome shortage.
Connect this elongation precision through the post-translational modification systems to understand how nascent proteins achieve functional conformations.
⚖️ The Protein Assembly Cycle: Elongation and Termination Precision
🔗 The Protein Maturation Network: Co-translational and Post-translational Processing
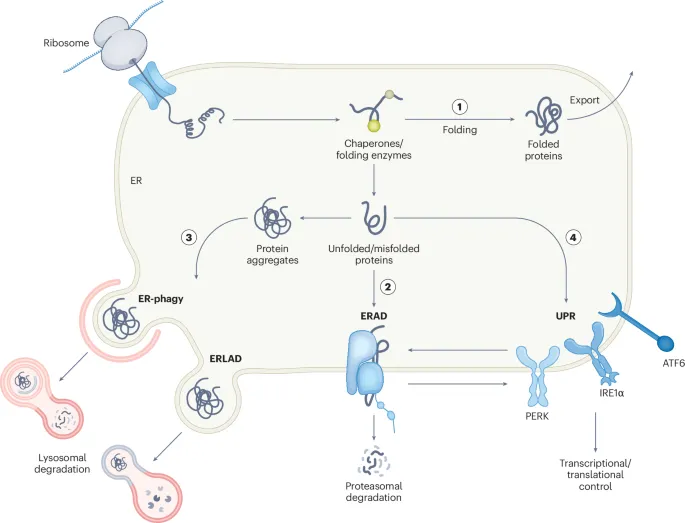
Nascent proteins undergo co-translational processing beginning during translation. Signal recognition particle (SRP) identifies ER signal sequences within 70-100 nucleotides of translation start, while cytosolic chaperones prevent misfolding of nascent chains.
📌 Remember: FOLD - Folding chaperones, Organellar targeting, Location signals, Degradation pathways
Hsp70 chaperones bind nascent polypeptides every 30-40 amino acids, preventing aggregation and facilitating folding. Hsp90 assists complex protein maturation, while chaperonins (GroEL/GroES in prokaryotes, CCT in eukaryotes) provide isolated folding chambers.
- Co-translational Targeting Pathways
- ER targeting: SRP recognition of hydrophobic signal sequences
- SRP54 binds signal peptides with nanomolar affinity
- SRP receptor docking occurs within seconds of ribosome binding
- Mitochondrial targeting: N-terminal presequences with positive charge
- TOM/TIM complexes coordinate import with ATP hydrolysis
- Matrix processing peptidase removes presequences after import
- ER targeting: SRP recognition of hydrophobic signal sequences
⭐ Clinical Pearl: α1-Antitrypsin deficiency results from Z mutation (Glu342Lys) causing protein misfolding, ER retention, and polymerization->85% of mutant protein undergoes ER-associated degradation.
| Modification Type | Timing | Frequency | Clinical Example | Therapeutic Target |
|---|---|---|---|---|
| Signal Cleavage | Co-translational | 30% of proteins | Preproinsulin | Signal peptidases |
| N-glycosylation | Co-translational | 50% of ER proteins | Congenital CDG | Glycosylation inhibitors |
| Phosphorylation | Post-translational | 30% of proteome | Kinase diseases | Kinase inhibitors |
| Ubiquitination | Post-translational | 80% turnover | Proteasome diseases | Proteasome inhibitors |
| SUMOylation | Post-translational | 10% of nuclear | Transcription control | SUMO inhibitors |
| %%{init: {'flowchart': {'htmlLabels': true}}}%% | ||||
| flowchart TD |
Start["🧬 Nascent Protein
• New polypeptide• Ribosome product"]
Signal{"❓ Signal Present?
• Sorting sequence• N-terminal tail"}
%% ER Pathway
SRP["🔌 SRP Recognition
• Signal particles• Translation pause"]
Trans["🕳️ ER Translocation
• Sec61 complex• Co-translational"]
QC["⚖️ ER Quality Control
• Glycan checking• Folding sensor"]
Folded{"✅ Properly Folded?
• Native state• Tertiary shape"}
Secretory["📦 Secretory Pathway
• Golgi transport• Exocytosis"]
ERAD["🗑️ ERAD Degradation
• Retro-translocation• Proteasome use"]
%% Mitochondrial Pathway
TOM["🔋 TOM/TIM Import
• Outer membrane• Inner membrane"]
MitoProc["⚙️ Mitochondrial Proc
• Cleave signal• Matrix folding"]
OrgFunc["⚡ Organellar Function
• ATP synthesis• Metabolic role"]
%% Cytosolic Pathway
CytoFold["💧 Cytosolic Folding
• Aqueous phase• Rapid process"]
Chap["🤝 Chaperone Assist
• HSP70 activity• Prevent clumps"]
CytoFunc["🛠️ Cytosolic Function
• Enzyme activity• Structural role"]
Start --> Signal Signal -->|ER Signal| SRP Signal -->|Mito Signal| TOM Signal -->|No Signal| CytoFold
SRP --> Trans --> QC --> Folded Folded -->|Yes| Secretory Folded -->|No| ERAD
TOM --> MitoProc --> OrgFunc CytoFold --> Chap --> CytoFunc
style Start fill:#F7F5FD, stroke:#F0EDFA, stroke-width:1.5px, rx:12, ry:12, color:#6B21A8 style Signal fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E style SRP fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C style Trans fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C style QC fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E style Folded fill:#FEF8EC, stroke:#FBECCA, stroke-width:1.5px, rx:12, ry:12, color:#854D0E style Secretory fill:#F1FCF5, stroke:#BEF4D8, stroke-width:1.5px, rx:12, ry:12, color:#166534 style ERAD fill:#FDF4F3, stroke:#FCE6E4, stroke-width:1.5px, rx:12, ry:12, color:#B91C1C style TOM fill:#FFF7ED, stroke:#FFEED5, stroke-width:1.5px, rx:12, ry:12, color:#C2410C style MitoProc fill:#EEFAFF, stroke:#DAF3FF, stroke-width:1.5px, rx:12, ry:12, color:#0369A1 style OrgFunc fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252 style CytoFold fill:#EEFAFF, stroke:#DAF3FF, stroke-width:1.5px, rx:12, ry:12, color:#0369A1 style Chap fill:#F1FCF5, stroke:#BEF4D8, stroke-width:1.5px, rx:12, ry:12, color:#166534 style CytoFunc fill:#F6F5F5, stroke:#E7E6E6, stroke-width:1.5px, rx:12, ry:12, color:#525252
> 💡 **Master This**: **Protein quality control networks** integrate **folding status** with **cellular stress responses**-**unfolded protein response** activation reduces **global translation** by **>70%** while **enhancing chaperone expression** by **5-10 fold**.
Connect this maturation network through the rapid clinical mastery tools to understand how transcription-translation knowledge transforms diagnostic and therapeutic decision-making.
🔗 The Protein Maturation Network: Co-translational and Post-translational Processing
🎯 Clinical Command Center: Transcription-Translation Mastery Arsenal
📌 Essential Numbers Arsenal: 50 nt/sec (RNA Pol speed), 20 aa/sec (ribosome rate), 99.99% (translation fidelity), 70S vs 80S (antibiotic selectivity), 5' cap (eukaryotic mRNA), 3 stop codons (UAG/UAA/UGA)
Rapid Diagnostic Patterns for transcription-translation disorders:
- Thalassemias: Globin gene mutations → Imbalanced chains → Hemolysis
- α-Thalassemia: Gene deletions on chromosome 16
- β-Thalassemia: Point mutations affecting transcription or splicing
- Ribosomopathies: Ribosomal protein defects → Growth retardation + Cancer predisposition
- Diamond-Blackfan: RPS19 mutations in 25% of cases
- Shwachman-Diamond: SBDS mutations affecting ribosome maturation
⭐ Clinical Pearl: Nonsense-mediated decay eliminates >90% of transcripts with premature stop codons, explaining why nonsense mutations often cause loss-of-function rather than dominant-negative effects.
| Clinical Scenario | Molecular Defect | Diagnostic Test | Treatment Approach | Prognosis Modifier |
|---|---|---|---|---|
| β-Thalassemia Major | β-globin transcription | Hb electrophoresis | Transfusion + chelation | HbF levels |
| Diamond-Blackfan Anemia | Ribosomal biogenesis | Ribosomal proteins | Corticosteroids | eADA activity |
| Vanishing White Matter | eIF2B mutations | MRI + genetics | Supportive care | Stress avoidance |
| Congenital CDG | N-glycosylation | Transferrin IEF | Mannose supplementation | PMM2 activity |
| Mitochondrial Disease | mt-tRNA mutations | Muscle biopsy | CoQ10 + vitamins | Heteroplasmy level |
Therapeutic Decision Framework:
- Transcriptional targets: RNA polymerase inhibitors (rifampin, α-amanitin)
- Translational targets: Ribosome inhibitors with species selectivity
- Processing targets: Splicing modulators for exon skipping therapy
- Quality control targets: Proteasome inhibitors for cancer treatment
This clinical mastery framework transforms molecular knowledge into diagnostic precision and therapeutic success, enabling evidence-based decisions that improve patient outcomes through mechanistic understanding.
🎯 Clinical Command Center: Transcription-Translation Mastery Arsenal
Practice Questions: Transcription/translation
Test your understanding with these related questions
A codon is an mRNA sequence consisting of 3 nucleotides that codes for an amino acid. Each position can be made up of any 4 nucleotides (A, U, G, C); therefore, there are a total of 64 (4 x 4 x 4) different codons that can be created but they only code for 20 amino acids. This is explained by the wobble phenomenon. One codon for leucine is CUU, which of the following can be another codon coding for leucine?












