DNA replication US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for DNA replication. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
DNA replication US Medical PG Question 1: A 71-year-old man with colorectal cancer comes to the physician for follow-up examination after undergoing a sigmoid colectomy. The physician recommends adjuvant chemotherapy with an agent that results in single-stranded DNA breaks. This chemotherapeutic agent most likely has an effect on which of the following enzymes?
- A. DNA polymerase III
- B. Topoisomerase I (Correct Answer)
- C. Helicase
- D. Telomerase
- E. Topoisomerase II
DNA replication Explanation: ***Topoisomerase I***
- **Topoisomerase I** creates **single-stranded DNA (ssDNA) breaks** to relieve torsional stress during DNA replication and transcription.
- Many chemotherapeutic agents, such as camptothecin and its derivatives (e.g., irinotecan, topotecan), target topoisomerase I, leading to DNA damage and apoptosis in cancer cells.
*DNA polymerase III*
- **DNA polymerase III** is primarily involved in bacterial DNA replication, synthesizing new DNA strands in a 5' to 3' direction.
- While essential for bacterial survival, it is not the target of chemotherapeutic agents that induce single-stranded DNA breaks in human cells.
*Helicase*
- **Helicase** is responsible for unwinding the DNA double helix during replication and transcription, separating the two strands.
- While its function is critical for DNA processes, it does not directly create DNA breaks as its primary mechanism of action.
*Telomerase*
- **Telomerase** is an enzyme that maintains telomere length at the ends of chromosomes, particularly active in cancer cells.
- Inhibitors of telomerase aim to shorten telomeres, leading to cellular senescence or apoptosis, but they do not primarily cause single-stranded DNA breaks.
*Topoisomerase II*
- **Topoisomerase II** creates **double-stranded DNA (dsDNA) breaks** to untangle and decatenate DNA.
- Though also a target for chemotherapy (e.g., etoposide, doxorubicin), its mechanism involves double-stranded breaks, not single-stranded breaks as specified in the question.
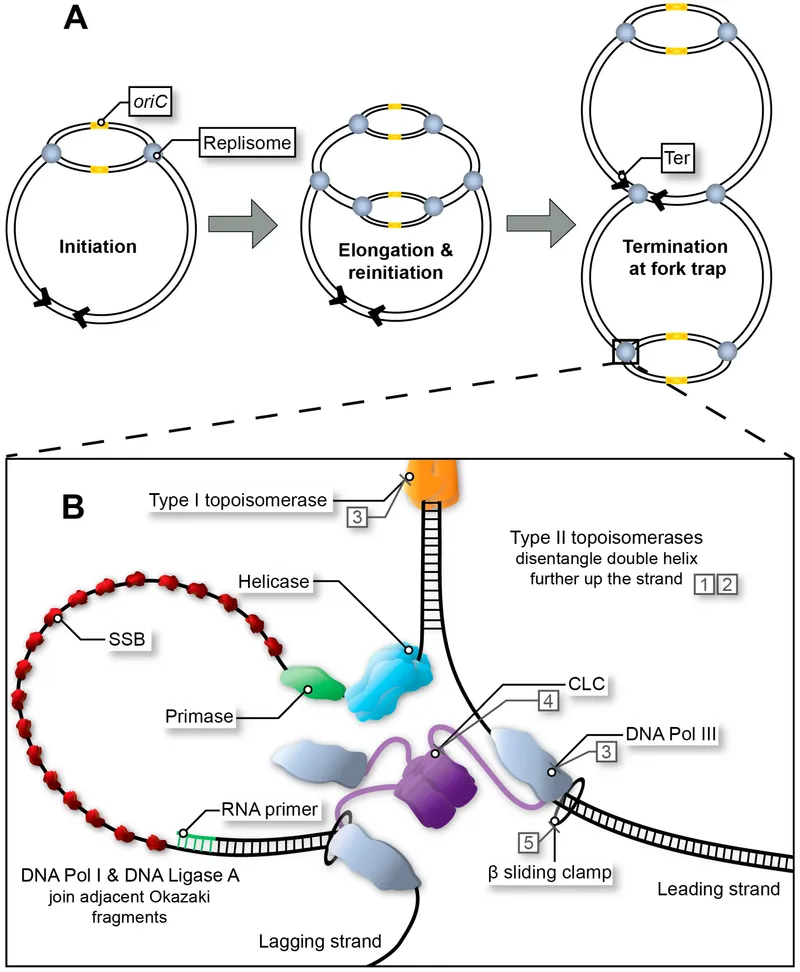
DNA replication US Medical PG Question 2: An investigator isolates bacteria from a patient who presented with dysuria and urinary frequency. These bacteria grow rapidly in pink colonies on MacConkey agar. During replication of these bacteria, the DNA strands are unwound at the origin of replication, forming two Y-shaped replication forks that open in opposite directions. At each replication fork, daughter strands are synthesized from the template strands in a 5′ to 3′ direction. On one strand, the DNA is synthesized continuously; on the other strand, the DNA is synthesized in short segments. The investigator finds that three enzymes are directly involved in elongating the DNA of the lagging strand in these bacteria. One of these enzymes has an additional function that the others do not possess. Which of the following steps in DNA replication is unique to this enzyme?
- A. Elongation of lagging strand in 5'→3' direction
- B. Excision of nucleotides with 5'→3' exonuclease activity (Correct Answer)
- C. Prevention of reannealing of the leading strand and the lagging strand
- D. Creation of ribonucleotide primers
- E. Proofreading for mismatched nucleotides
DNA replication Explanation: ***Excision of nucleotides with 5'→3' exonuclease activity***
- **DNA polymerase I** possesses unique **5'→3' exonuclease activity** that allows it to remove RNA primers synthesized by primase.
- After primer removal, DNA polymerase I synthesizes DNA in the 5'→3' direction to fill the gap.
*Elongation of lagging strand in 5'→3' direction*
- While **DNA polymerase I** elongates the lagging strand, this 5'→3' synthesis function is also shared by **DNA polymerase III**, which is the primary enzyme for DNA synthesis.
- Therefore, this specific function is not unique to the enzyme in question (DNA polymerase I) as DNA polymerase III also performs 5'→3' elongation.
*Prevention of reannealing of the leading strand and the lagging strand*
- This function is carried out by **single-strand binding proteins (SSBs)**, which bind to the separated DNA strands to prevent them from reannealing and protect them from degradation.
- This is not a function of any DNA polymerase.
*Creation of ribonucleotide primers*
- The synthesis of short RNA primers required for initiation of DNA synthesis is performed by **primase**, an RNA polymerase.
- DNA polymerases do not create primers but rather extend them.
*Proofreading for mismatched nucleotides*
- **DNA polymerase I** and **DNA polymerase III** both possess **3'→5' exonuclease activity** for proofreading, allowing them to remove incorrectly incorporated nucleotides.
- Since this function is shared by DNA polymerase III, it is not unique to DNA polymerase I.
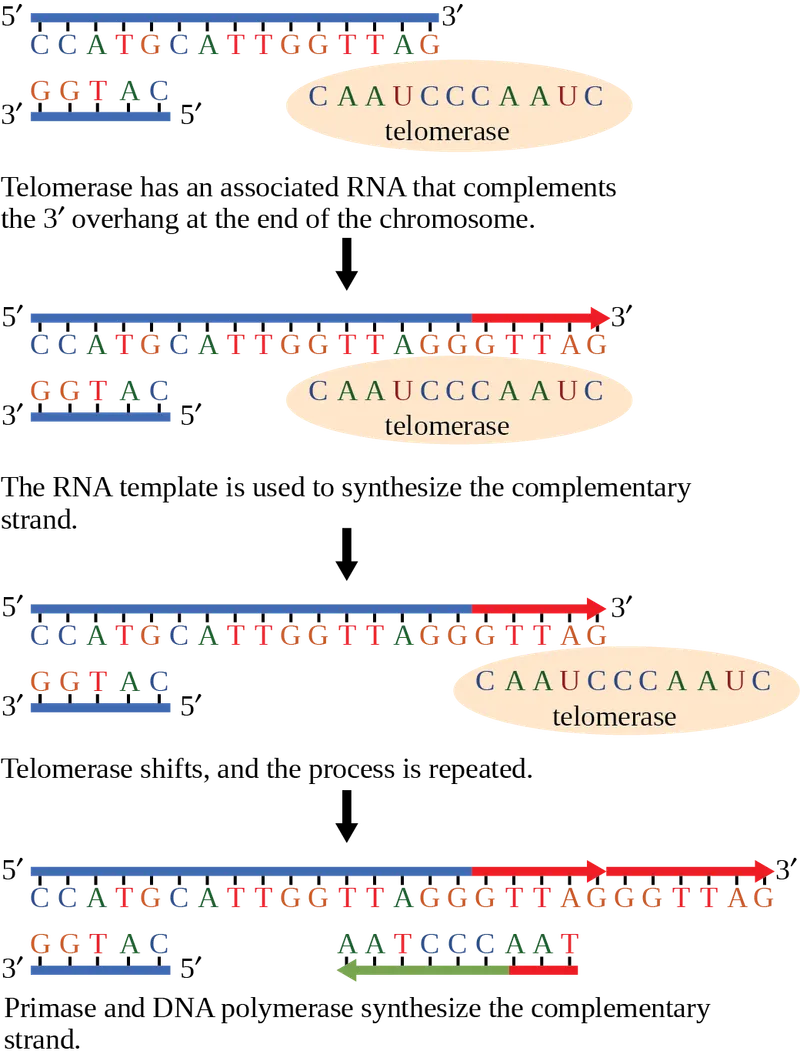
DNA replication US Medical PG Question 3: An investigator is studying the biology of human sperm cells. She isolates spermatogonia obtained on a testicular biopsy from a group of healthy male volunteers. She finds that the DNA of spermatogonia obtained from these men show a large number of TTAGGG sequence repeats. This finding can best be explained by increased activity of an enzyme with which of the following functions?
- A. Ligation of Okazaki fragments
- B. Proofreading of synthesized daughter strands
- C. RNA-dependent synthesis of DNA (Correct Answer)
- D. Production of short RNA sequences
- E. Hemimethylation of DNA strand
DNA replication Explanation: ***RNA-dependent synthesis of DNA***
- The TTAGGG sequence repeats are **telomeric sequences**, which are maintained by **telomerase**, an enzyme that synthesizes DNA from an RNA template.
- **Spermatogonia** are germline stem cells that express high levels of telomerase to maintain telomere length across generations.
*Ligation of Okazaki fragments*
- This function is carried out by **DNA ligase**, which joins discontinuous DNA fragments during replication on the lagging strand.
- This process is essential for general DNA replication but is not specific to the formation or maintenance of telomeric repeats.
*Proofreading of synthesized daughter strands*
- This is a function of **DNA polymerase exonuclease activity**, which corrects errors during DNA replication.
- While important for genetic fidelity, it does not explain the presence or increase of specific TTAGGG repeat sequences at telomeres.
*Production of short RNA sequences*
- This function is performed by **primase**, which synthesizes RNA primers necessary to initiate DNA synthesis during replication.
- These RNA primers are later removed and replaced with DNA, and this process is not directly responsible for generating or extending telomeric repeats.
*Hemimethylation of DNA strand*
- Hemimethylation occurs during **DNA replication** when new DNA strands are unmethylated while parental strands are methylated.
- This phenomenon is involved in DNA repair and gene regulation but is unrelated to the synthesis or regulation of telomeric sequences.
DNA replication US Medical PG Question 4: A laboratory physician investigates the chromosomes of a fetus with a suspected chromosomal anomaly. She processes a cell culture obtained by amniocentesis. Prior to staining and microscopic examination of the fetal chromosomes, a drug that blocks cell division is added to the cell culture. In order to arrest chromosomes in metaphase, the physician most likely added a drug that is also used for the treatment of which of the following conditions?
- A. Trichomonas vaginitis
- B. Testicular cancer
- C. Herpes zoster
- D. Polycythemia vera
- E. Acute gouty arthritis (Correct Answer)
DNA replication Explanation: ***Acute gouty arthritis***
- The drug used to arrest chromosomes in metaphase is likely **colchicine**, which inhibits **microtubule polymerization** and spindle formation, thus arresting cells in metaphase.
- **Colchicine** is a well-established treatment for **acute gouty arthritis** due to its anti-inflammatory properties, primarily through disrupting neutrophil functions.
*Trichomonas vaginitis*
- This condition is typically treated with **metronidazole** or **tinidazole**, which are antibiotics targeting protozoa and anaerobic bacteria.
- These drugs do not inhibit microtubule assembly or arrest cells in metaphase.
*Testicular cancer*
- Testicular cancer is primarily treated with **BEP regimen** (bleomycin, etoposide, cisplatin), which does not include microtubule-inhibiting agents.
- While vinca alkaloids (vincristine, vinblastine) do arrest cells in metaphase via microtubule inhibition similar to colchicine, they are not standard first-line agents for testicular cancer.
- The question specifically asks about the primary clinical use of colchicine, which is gout, not cancer chemotherapy.
*Herpes zoster*
- Herpes zoster (shingles) is a viral infection treated with **antiviral medications** like acyclovir, valacyclovir, or famciclovir.
- These antivirals work by interfering with viral DNA replication and do not target microtubule formation or cell division.
*Polycythemia vera*
- Polycythemia vera is a myeloproliferative neoplasm often managed with **phlebotomy**, **hydroxyurea**, or ruxolitinib.
- These treatments aim to reduce blood cell counts or inhibit specific signaling pathways, none of which primarily involve arresting cells in metaphase by disrupting microtubules.
DNA replication US Medical PG Question 5: A mutant stem cell was created by using an inducible RNAi system, such that when doxycycline is added, the siRNA targeting DNA helicase is expressed, effectively knocking down the gene for DNA helicase. Which of the following will occur during DNA replication?
- A. The RNA primer is not created
- B. DNA is not unwound (Correct Answer)
- C. The two melted DNA strands reanneal
- D. DNA supercoiling is not relieved
- E. Newly synthesized DNA fragments are not ligated
DNA replication Explanation: ***DNA is not unwound***
- **DNA helicase** is essential for unwinding the **double-stranded DNA** helix, separating it into two single strands. This process creates the **replication fork**.
- Without functional DNA helicase due to **gene knockdown**, the DNA helix cannot be unwound, thus halting DNA replication.
*The RNA primer is not created*
- **RNA primers** are synthesized by **primase**, an enzyme distinct from DNA helicase.
- While unwinding is necessary for primer synthesis, the *creation* of the primer itself is a function of primase.
*The two melted DNA strands reanneal*
- **Reannealing** of DNA strands is prevented by **single-strand binding proteins (SSBs)**, which bind to the separated single strands.
- While helicase unwinds, SSBs specifically keep the strands apart to allow DNA polymerase access.
*DNA supercoiling is not relieved*
- **DNA supercoiling** is relieved by **topoisomerases**, enzymes that cut, unwind, and religate DNA strands to reduce torsional stress.
- This is a distinct function from DNA helicase, which focuses on breaking hydrogen bonds between strands.
*Newly synthesized DNA fragments are not ligated*
- **Ligation** of newly synthesized **Okazaki fragments** on the lagging strand is performed by **DNA ligase**.
- This process occurs downstream from the unwinding step facilitated by DNA helicase.
DNA replication US Medical PG Question 6: As part of a clinical research study, the characteristics of neoplastic and normal cells are being analyzed in culture. It is observed that neoplastic cell division is aided by an enzyme which repairs progressive chromosomal shortening, which is not the case in normal cells. Due to the lack of chromosomal shortening, these neoplastic cells divide more rapidly than the normal cells. Which of the following enzymes is most likely involved?
- A. Topoisomerase
- B. DNA polymerase
- C. Reverse transcriptase
- D. Protein kinase
- E. Telomerase (Correct Answer)
DNA replication Explanation: ***Telomerase***
- **Telomerase** is an enzyme that adds repetitive nucleotide sequences (telomeres) to the ends of chromosomes, counteracting their progressive shortening during DNA replication. This activity is crucial for the continuous division of neoplastic cells.
- In normal somatic cells, **telomerase activity is typically low or absent**, leading to telomere shortening with each division, eventually triggering cellular senescence or apoptosis. The presence of telomerase in neoplastic cells allows them to bypass these natural limits on proliferation.
*Topoisomerase*
- **Topoisomerases** are enzymes that regulate the supercoiling of DNA by breaking and rejoining DNA strands, which is essential during replication and transcription to relieve torsional stress.
- They do not directly repair chromosomal shortening but rather manage the topological state of DNA.
*DNA polymerase*
- **DNA polymerase** is primarily responsible for synthesizing new DNA strands by adding nucleotides, thereby elongating the DNA molecule during replication and DNA repair processes.
- While essential for DNA replication, it cannot fully replicate the very ends of linear chromosomes, leading to the **end-replication problem** and telomere shortening.
*Reverse transcriptase*
- **Reverse transcriptase** is an enzyme that synthesizes DNA from an RNA template, a process central to retroviruses and some eukaryotic elements like retrotransposons.
- Although telomerase itself is a specialized reverse transcriptase (using an RNA template to synthesize DNA telomeres), the general term "reverse transcriptase" does not specifically refer to the enzyme that repairs chromosomal shortening in the context of cell division.
*Protein kinase*
- **Protein kinases** are enzymes that add phosphate groups to proteins, a process known as phosphorylation. This modification can alter protein activity, localization, or stability, playing a critical role in signal transduction pathways.
- They are involved in regulating various cellular processes, including cell growth and division, but do not directly repair chromosomal shortening.
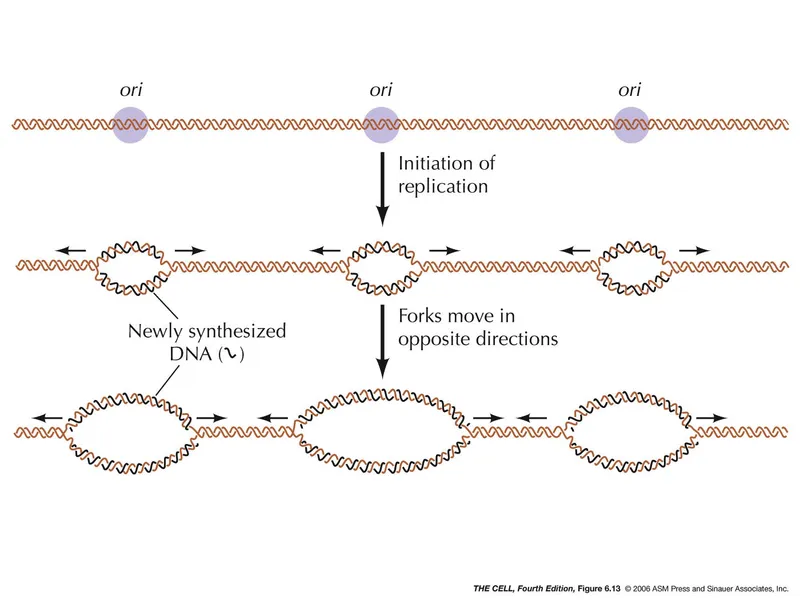
DNA replication US Medical PG Question 7: Although nucleotide addition during DNA replication in prokaryotes proceeds approximately 20-times faster than in eukaryotes, why can much larger amounts of DNA be replicated in eukaryotes in a time-effective manner?
- A. Eukaryotes have multiple origins of replication (Correct Answer)
- B. Eukaryotes have helicase which can more easily unwind DNA strands
- C. Eukaryotes have fewer polymerase types
- D. Eukaryotes have less genetic material to replicate
- E. Eukaryotes have a single, circular chromosome
DNA replication Explanation: ***Eukaryotes have multiple origins of replication***
- Eukaryotic chromosomes are much larger than prokaryotic chromosomes and require multiple origins of replication to complete DNA synthesis within a reasonable timeframe.
- Each origin of replication initiates simultaneously, allowing DNA synthesis to occur at many sites along the chromosome, effectively increasing the overall speed of replication.
- This compensates for the slower rate of nucleotide addition by DNA polymerase in eukaryotes compared to prokaryotes.
*Eukaryotes have helicase which can more easily unwind DNA strands*
- While helicase activity is crucial for unwinding DNA, there is no evidence to suggest that eukaryotic helicases are significantly more efficient or faster at unwinding DNA compared to prokaryotic helicases in a way that would account for the large difference in overall replication time.
- The rate of DNA unwinding by helicase is a factor in replication speed, but it does not overcome the fundamental limitation of a single origin of replication in prokaryotes.
*Eukaryotes have fewer polymerase types*
- Eukaryotic cells actually have **more** types of DNA polymerases than prokaryotic cells, each specialized for different functions like replication, repair, and mitochondrial DNA synthesis.
- The number of polymerase types does not directly relate to the speed or efficiency of overall DNA replication in terms of replicating large amounts of DNA.
*Eukaryotes have less genetic material to replicate*
- Eukaryotic organisms typically have significantly **more** genetic material (a larger genome size) than prokaryotic organisms, not less.
- If eukaryotes had less genetic material, the question itself about effective replication of "much larger amounts of DNA" would be contradictory.
*Eukaryotes have a single, circular chromosome*
- Eukaryotic cells have **multiple, linear chromosomes** within a membrane-bound nucleus, not a single circular chromosome.
- Prokaryotic cells typically have a single, circular chromosome located in the nucleoid region.
- The linear structure of eukaryotic chromosomes with multiple origins is actually what enables efficient replication of large genomes, making this statement both factually incorrect and contradictory to the mechanism in question.
DNA replication US Medical PG Question 8: A 12-year-old girl is brought to an oncologist, as she was recently diagnosed with a rare form of cancer. Cytogenetic studies reveal that the tumor is responsive to vinblastine, which is a cell-cycle specific anticancer agent. It acts on the M phase of the cell cycle and inhibits the growth of cells. Which of the following statements best describes the regulation of the cell cycle?
- A. Inhibitors of DNA synthesis act in the M phase of the cell cycle.
- B. The G0 phase is the checkpoint before G1.
- C. Cyclin-dependent activation of CDK1 (CDC2) takes place upon the entry of a cell into M phase of the cell cycle. (Correct Answer)
- D. EGF from a blood clot stimulates the growth and proliferation of cells in the healing process.
- E. Replication of the genome occurs in the M phase of the cell cycle.
DNA replication Explanation: ***Cyclin-dependent activation of CDK1 (CDC2) takes place upon the entry of a cell into M phase of the cell cycle.***
- The **M-phase promoting factor (MPF)**, composed of **CDK1 (CDC2)** and **cyclin B**, is activated at the G2/M transition, driving the cell into mitosis.
- Activation of CDK1 by **cyclin B binding** and subsequent dephosphorylation of threonine 161 is crucial for initiation of mitosis.
*Inhibitors of DNA synthesis act in the M phase of the cell cycle.*
- **Inhibitors of DNA synthesis**, such as **hydroxyurea** and **methotrexate**, primarily act during the **S phase** of the cell cycle, when DNA replication occurs.
- The M phase is characterized by **mitosis** (nuclear division) and **cytokinesis** (cytoplasmic division), not DNA synthesis.
*The G0 phase is the checkpoint before G1.*
- The **G0 phase** is a **resting state** where cells exit the cell cycle and cease to divide, not a checkpoint before G1.
- The main checkpoint before G1 is typically referred to as the **restriction point** or **G1 checkpoint**, which determines if a cell will commit to division.
*EGF from a blood clot stimulates the growth and proliferation of cells in the healing process.*
- While **EGF (Epidermal Growth Factor)** does stimulate cell growth and proliferation in healing, it is not primarily associated with blood clots.
- **Platelets** in blood clots release growth factors like **PDGF (Platelet-Derived Growth Factor)** and **TGF-β (Transforming Growth Factor-beta)**, which are critical for wound healing.
*Replication of the genome occurs in the M phase of the cell cycle.*
- **Replication of the genome** (DNA synthesis) occurs during the **S phase** (synthesis phase) of the cell cycle.
- The **M phase** is dedicated to **mitosis** (separation of duplicated chromosomes) and **cytokinesis**, where the cell divides into two daughter cells.
DNA replication US Medical PG Question 9: An investigator studying DNA replication in Campylobacter jejuni inoculates a strain of this organism into a growth medium that contains radiolabeled thymine. After 2 hours, the rate of incorporation of radiolabeled thymine is measured as a proxy for the rate of DNA replication. The cells are then collected by centrifugation and suspended in a new growth medium that lacks ribonucleotides. After another 2 hours, the rate of incorporation of radiolabeled thymine is measured again. The new growth medium directly affects the function of which of the following enzymes?
- A. DNA polymerase II
- B. Telomerase
- C. Primase (Correct Answer)
- D. DNA polymerase I
- E. Ligase
DNA replication Explanation: ***Primase***
- **Primase** is an **RNA polymerase** that synthesizes short **RNA primers** required for DNA replication. The new growth medium lacks **ribonucleotides**, which are the building blocks for RNA.
- Without **ribonucleotides**, primase cannot synthesize RNA primers, thereby directly affecting its function and subsequently inhibiting DNA replication.
*DNA polymerase II*
- **DNA polymerase II** is primarily involved in **DNA repair** and translesion synthesis, not in synthesizing the main leading and lagging strands of DNA replication.
- Its function is not directly dependent on the availability of **ribonucleotides** for primer synthesis during normal replication.
*DNA polymerase I*
- **DNA polymerase I** is crucial for removing **RNA primers** and filling in the resulting gaps with DNA nucleotides.
- While it acts on the primers made by primase, its direct catalytic activity does not involve synthesizing RNA primers from **ribonucleotides**.
*Telomerase*
- **Telomerase** is a specialized reverse transcriptase that extends telomeres at the ends of eukaryotic chromosomes.
- **Campylobacter jejuni** is a prokaryote and therefore lacks linear chromosomes and **telomeres**, making telomerase irrelevant to its DNA replication.
*Ligase*
- **Ligase** is an enzyme that joins **Okazaki fragments** and other DNA breaks by forming phosphodiester bonds.
- Its function involves sealing nicks in the DNA backbone and does not directly rely on the presence of **ribonucleotides** for creating new primers.
DNA replication US Medical PG Question 10: Replication in eukaryotic cells is a highly organized and accurate process. The process involves a number of enzymes such as primase, DNA polymerase, topoisomerase II, and DNA ligase. In which of the following directions is DNA newly synthesized?
- A. 3' --> 5'
- B. N terminus --> C terminus
- C. C terminus --> N terminus
- D. 3' --> 5' & 5' --> 3'
- E. 5' --> 3' (Correct Answer)
DNA replication Explanation: ***5' --> 3'***
- DNA polymerase can only add **nucleotides** to the 3' end of a growing strand, meaning synthesis always proceeds in a **5' to 3' direction**.
- This is true for both the **leading strand** (synthesized continuously) and the **lagging strand** (synthesized discontinuously via Okazaki fragments).
*3' --> 5'*
- While the parental template strand is read in the 3' to 5' direction, the *newly synthesized* DNA strand is always built in the **opposite, antiparallel 5' to 3' direction**.
- DNA polymerase lacks the ability to add new nucleotides to the **5' phosphate group** of the growing strand.
*N terminus --> C terminus*
- This directional notation refers to the synthesis of **proteins**, where amino acids are added to the C (carboxyl) terminus of the growing polypeptide chain.
- It does not apply to the synthesis direction of **nucleic acids (DNA or RNA)**.
*C terminus --> N terminus*
- This directional notation is incorrectly applied; protein synthesis always proceeds from the **N (amino) terminus to the C (carboxyl) terminus**.
- This has no relevance to the synthesis direction of **DNA**.
*3' --> 5' & 5' --> 3'*
- Although DNA replication involves two strands, one is synthesized continuously in the **5' → 3' direction (leading strand)** and the other discontinuously, but still *each fragment* is synthesized in the **5' → 3' direction (lagging strand)**.
- No new DNA strand is synthesized in the **3' → 5' direction**.
More DNA replication US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.