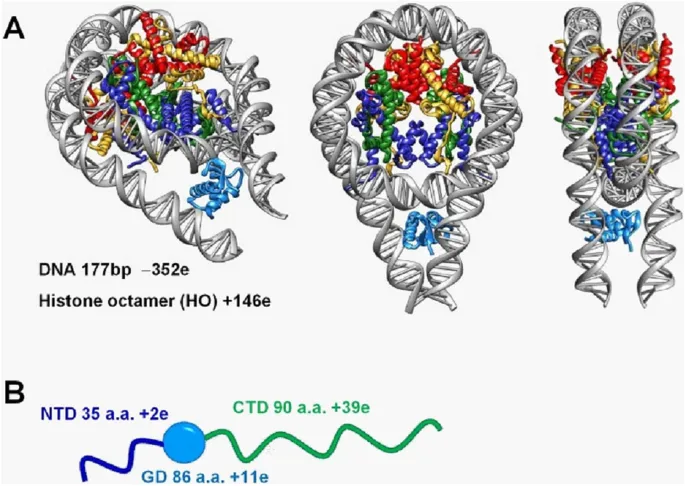
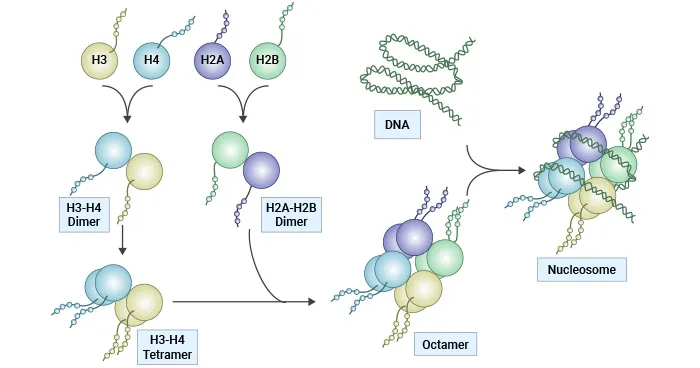
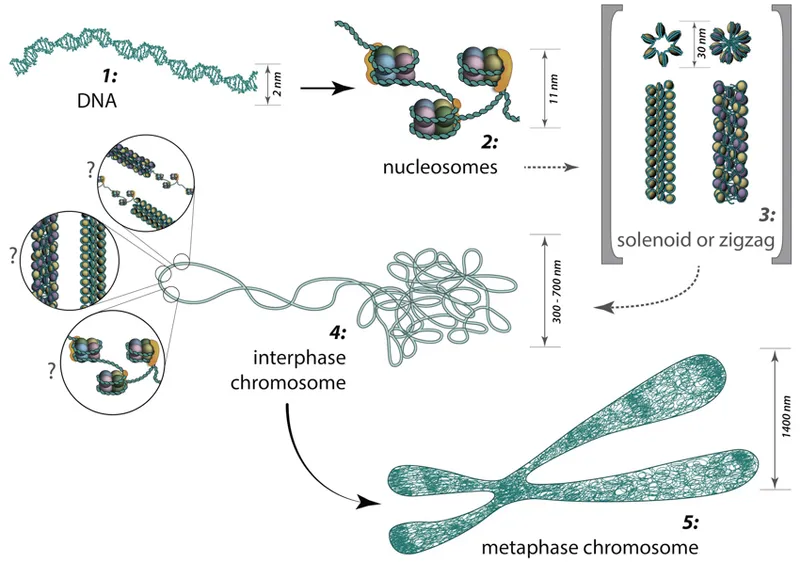
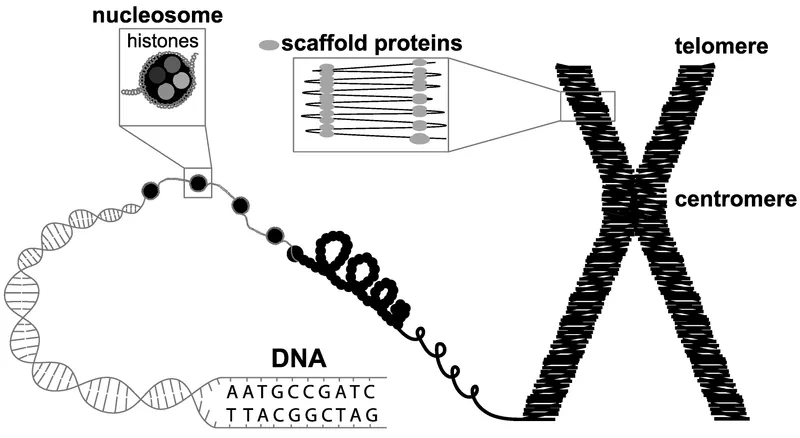
Chromosomal structure US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Chromosomal structure. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Chromosomal structure US Medical PG Question 1: A researcher is investigating compounds that modulate the cell cycle as possible chemotherapeutic agents against peripheral T-cell lymphoma. The researcher discovers a group of natural compounds with inhibitory activity against histone deacetylases, a class of enzymes that remove acetyl groups from the lysine residues of histones. A histone deacetylase inhibitor most likely causes which of the following?
- A. Prevention of DNA strand reannealing
- B. Increased heterochromatin formation
- C. Suppression of gene transcription
- D. Relaxation of DNA coiling (Correct Answer)
- E. Tighter coiling of DNA
Chromosomal structure Explanation: ***Relaxation of DNA coiling***
- Histone deacetylase (HDAC) inhibitors block the removal of **acetyl groups** from **histones**, leading to increased histone acetylation.
- Increased acetylation **reduces the positive charge** of histones, loosening their grip on the negatively charged DNA and causing **relaxation of DNA coiling**.
*Prevention of DNA strand reannealing*
- This process is primarily influenced by factors affecting **hydrogen bonding** between DNA strands, such as **temperature** or **DNA denaturing agents**, not directly by histone acetylation.
- DNA reannealing is the reformation of a **double helix** from single strands, a different mechanism than chromatin structure.
*Increased heterochromatin formation*
- **Heterochromatin** is characterized by **tightly coiled DNA** and is associated with **deacetylated histones** and gene silencing.
- Increased acetylation, as caused by HDAC inhibitors, would lead to less heterochromatin and more **euchromatin**.
*Suppression of gene transcription*
- **Relaxation of DNA coiling** makes the DNA more accessible to transcription factors and RNA polymerase, thereby generally **promoting gene transcription**, not suppressing it.
- **HDAC inhibitors** primarily promote gene expression by increasing the accessibility of DNA to the transcriptional machinery.
*Tighter coiling of DNA*
- **Deacetylation of histones** leads to stronger interaction between histones and DNA, resulting in **tighter coiling** and chromatin condensation.
- HDAC inhibitors, by preventing deacetylation, promote the opposite effect: **DNA uncoiling** and relaxation.
Chromosomal structure US Medical PG Question 2: An investigator is studying the biology of human sperm cells. She isolates spermatogonia obtained on a testicular biopsy from a group of healthy male volunteers. She finds that the DNA of spermatogonia obtained from these men show a large number of TTAGGG sequence repeats. This finding can best be explained by increased activity of an enzyme with which of the following functions?
- A. Ligation of Okazaki fragments
- B. Proofreading of synthesized daughter strands
- C. RNA-dependent synthesis of DNA (Correct Answer)
- D. Production of short RNA sequences
- E. Hemimethylation of DNA strand
Chromosomal structure Explanation: ***RNA-dependent synthesis of DNA***
- The TTAGGG sequence repeats are **telomeric sequences**, which are maintained by **telomerase**, an enzyme that synthesizes DNA from an RNA template.
- **Spermatogonia** are germline stem cells that express high levels of telomerase to maintain telomere length across generations.
*Ligation of Okazaki fragments*
- This function is carried out by **DNA ligase**, which joins discontinuous DNA fragments during replication on the lagging strand.
- This process is essential for general DNA replication but is not specific to the formation or maintenance of telomeric repeats.
*Proofreading of synthesized daughter strands*
- This is a function of **DNA polymerase exonuclease activity**, which corrects errors during DNA replication.
- While important for genetic fidelity, it does not explain the presence or increase of specific TTAGGG repeat sequences at telomeres.
*Production of short RNA sequences*
- This function is performed by **primase**, which synthesizes RNA primers necessary to initiate DNA synthesis during replication.
- These RNA primers are later removed and replaced with DNA, and this process is not directly responsible for generating or extending telomeric repeats.
*Hemimethylation of DNA strand*
- Hemimethylation occurs during **DNA replication** when new DNA strands are unmethylated while parental strands are methylated.
- This phenomenon is involved in DNA repair and gene regulation but is unrelated to the synthesis or regulation of telomeric sequences.
Chromosomal structure US Medical PG Question 3: A group of scientists developed a mouse model to study nondisjunction in meiosis. Their mouse model produced gametes in the following ratio: 2 gametes with 24 chromosomes each and 2 gametes with 22 chromosomes each. In which of the following steps of meiosis did the nondisjunction occur?
- A. Telophase I
- B. Metaphase II
- C. Anaphase I (Correct Answer)
- D. Anaphase II
- E. Metaphase I
Chromosomal structure Explanation: ***Anaphase I***
- Nondisjunction during **Anaphase I** occurs when homologous chromosomes fail to separate properly, meaning both homologs of a chromosome pair go to the same pole.
- This results in two secondary gametocytes with abnormal chromosome numbers: one with n+1 chromosomes (24) and one with n-1 chromosomes (22).
- When meiosis II proceeds normally, each abnormal secondary gametocyte divides to produce 2 identical gametes, resulting in **all 4 gametes being abnormal** in a 2:2 ratio (two n+1 and two n-1), matching the observed pattern.
*Telophase I*
- **Telophase I** is the final stage of meiosis I where chromosomes arrive at the poles and the cell divides, but it's not where the initial separation error (nondisjunction) occurs.
- Nondisjunction happens due to a failure of **chromosome segregation**, which is a process of anaphase, not telophase.
*Metaphase II*
- **Metaphase II** involves the alignment of sister chromatids at the metaphase plate in secondary gametocytes. Nondisjunction at this stage would involve sister chromatids failing to separate.
- Nondisjunction in Metaphase II (or Anaphase II) would lead to 2 normal gametes (23 chromosomes), one gamete with n+1 (24 chromosomes), and one gamete with n-1 (22 chromosomes), which differs from the given ratio.
*Anaphase II*
- **Nondisjunction in Anaphase II** would involve the failure of sister chromatids to separate in one of the secondary gametocytes.
- This would produce two normal gametes (23 chromosomes), one gamete with 24 chromosomes (n+1), and one gamete with 22 chromosomes (n-1), which is not the 2:2 ratio observed.
*Metaphase I*
- **Metaphase I** is characterized by the alignment of homologous chromosome pairs at the metaphase plate. While an issue here could precede nondisjunction, the actual event of failed separation occurs during anaphase.
- No separation of chromosomes occurs in Metaphase I; it is the stage of **chromosome alignment** before segregation.
Chromosomal structure US Medical PG Question 4: Replication in eukaryotic cells is a highly organized and accurate process. The process involves a number of enzymes such as primase, DNA polymerase, topoisomerase II, and DNA ligase. In which of the following directions is DNA newly synthesized?
- A. 3' --> 5'
- B. N terminus --> C terminus
- C. C terminus --> N terminus
- D. 3' --> 5' & 5' --> 3'
- E. 5' --> 3' (Correct Answer)
Chromosomal structure Explanation: ***5' --> 3'***
- DNA polymerase can only add **nucleotides** to the 3' end of a growing strand, meaning synthesis always proceeds in a **5' to 3' direction**.
- This is true for both the **leading strand** (synthesized continuously) and the **lagging strand** (synthesized discontinuously via Okazaki fragments).
*3' --> 5'*
- While the parental template strand is read in the 3' to 5' direction, the *newly synthesized* DNA strand is always built in the **opposite, antiparallel 5' to 3' direction**.
- DNA polymerase lacks the ability to add new nucleotides to the **5' phosphate group** of the growing strand.
*N terminus --> C terminus*
- This directional notation refers to the synthesis of **proteins**, where amino acids are added to the C (carboxyl) terminus of the growing polypeptide chain.
- It does not apply to the synthesis direction of **nucleic acids (DNA or RNA)**.
*C terminus --> N terminus*
- This directional notation is incorrectly applied; protein synthesis always proceeds from the **N (amino) terminus to the C (carboxyl) terminus**.
- This has no relevance to the synthesis direction of **DNA**.
*3' --> 5' & 5' --> 3'*
- Although DNA replication involves two strands, one is synthesized continuously in the **5' → 3' direction (leading strand)** and the other discontinuously, but still *each fragment* is synthesized in the **5' → 3' direction (lagging strand)**.
- No new DNA strand is synthesized in the **3' → 5' direction**.
Chromosomal structure US Medical PG Question 5: A 62-year-old man with small cell lung cancer undergoes radiation therapy. His oncologist explains that radiation causes DNA damage and double strand breaks and this damage stops the cancer cells from growing because they can no longer replicate their DNA. One key mediator of this process is a cell cycle regulator called P53, which is upregulated after DNA damage and helps to trigger cell cycle arrest and apoptosis. One mechanism by which P53 activity is increased is a certain chromatin modification that loosens DNA coiling allowing for greater transcription of the proteins within that region of DNA. Which of the following enzymes most likely causes the chromatin modification described in this case?
- A. Histone deacetylase
- B. Histone acetyltransferase (Correct Answer)
- C. Histone methyltransferase
- D. DNA methyltransferase
- E. Xist
Chromosomal structure Explanation: ***Histone acetyltransferase***
- This enzyme **acetylates histone proteins**, neutralizing their positive charge and thereby weakening their interaction with negatively charged DNA.
- This modification leads to a more **relaxed chromatin structure (euchromatin)**, making DNA more accessible for **transcription**, which is consistent with the upregulation of P53.
*Histone deacetylase*
- This enzyme **removes acetyl groups from histones**, making them more positively charged and increasing their affinity for DNA.
- This results in **condensed chromatin (heterochromatin)**, which generally **represses gene transcription**.
*Histone methyltransferase*
- This enzyme **adds methyl groups to histones**, which can either activate or repress gene transcription depending on the specific **lysine or arginine residue** methylated and the number of methyl groups added.
- While methylation is a chromatin modification, the question specifically describes a process of **loosening DNA coiling for greater transcription**, which is more characteristic of acetylation.
*DNA methyltransferase*
- This enzyme **adds methyl groups directly to DNA**, typically at **CpG sites**, leading to **gene silencing** by hindering transcription factor binding or recruiting repressor complexes.
- This modification primarily affects DNA directly, not histone proteins, and generally **inhibits gene expression**.
*Xist*
- **Xist (X-inactive specific transcript)** is a **long non-coding RNA** that plays a crucial role in **X-chromosome inactivation** in females.
- It functions by coating one of the X chromosomes, leading to its transcriptional silencing, rather than directly modifying chromatin for general gene upregulation.
Chromosomal structure US Medical PG Question 6: As part of a clinical research study, the characteristics of neoplastic and normal cells are being analyzed in culture. It is observed that neoplastic cell division is aided by an enzyme which repairs progressive chromosomal shortening, which is not the case in normal cells. Due to the lack of chromosomal shortening, these neoplastic cells divide more rapidly than the normal cells. Which of the following enzymes is most likely involved?
- A. Topoisomerase
- B. DNA polymerase
- C. Reverse transcriptase
- D. Protein kinase
- E. Telomerase (Correct Answer)
Chromosomal structure Explanation: ***Telomerase***
- **Telomerase** is an enzyme that adds repetitive nucleotide sequences (telomeres) to the ends of chromosomes, counteracting their progressive shortening during DNA replication. This activity is crucial for the continuous division of neoplastic cells.
- In normal somatic cells, **telomerase activity is typically low or absent**, leading to telomere shortening with each division, eventually triggering cellular senescence or apoptosis. The presence of telomerase in neoplastic cells allows them to bypass these natural limits on proliferation.
*Topoisomerase*
- **Topoisomerases** are enzymes that regulate the supercoiling of DNA by breaking and rejoining DNA strands, which is essential during replication and transcription to relieve torsional stress.
- They do not directly repair chromosomal shortening but rather manage the topological state of DNA.
*DNA polymerase*
- **DNA polymerase** is primarily responsible for synthesizing new DNA strands by adding nucleotides, thereby elongating the DNA molecule during replication and DNA repair processes.
- While essential for DNA replication, it cannot fully replicate the very ends of linear chromosomes, leading to the **end-replication problem** and telomere shortening.
*Reverse transcriptase*
- **Reverse transcriptase** is an enzyme that synthesizes DNA from an RNA template, a process central to retroviruses and some eukaryotic elements like retrotransposons.
- Although telomerase itself is a specialized reverse transcriptase (using an RNA template to synthesize DNA telomeres), the general term "reverse transcriptase" does not specifically refer to the enzyme that repairs chromosomal shortening in the context of cell division.
*Protein kinase*
- **Protein kinases** are enzymes that add phosphate groups to proteins, a process known as phosphorylation. This modification can alter protein activity, localization, or stability, playing a critical role in signal transduction pathways.
- They are involved in regulating various cellular processes, including cell growth and division, but do not directly repair chromosomal shortening.
Chromosomal structure US Medical PG Question 7: A 23-year-old man presents to his primary care physician with 2 weeks of headache, palpitations, and excessive sweating. He has no past medical history and his family history is significant for clear cell renal cell carcinoma in his father as well as retinal hemangioblastomas in his older sister. On presentation his temperature is 99°F (37.2°C), blood pressure is 181/124 mmHg, pulse is 105/min, and respirations are 18/min. After administration of appropriate medications, he is taken emergently for surgical removal of a mass that was detected by abdominal computed tomography scan. A mutation on which of the following chromosomes would most likely be seen in this patient?
- A. 3 (Correct Answer)
- B. 11
- C. 2
- D. 17
- E. 10
Chromosomal structure Explanation: ***Chromosome 3***
- The patient's symptoms (**headache, palpitations, excessive sweating**) and **hypertension** (181/124 mmHg, pulse 105/min) suggest a **pheochromocytoma**, which is a catecholamine-secreting tumor often found in the adrenal medulla. The abdominal CT finding of a mass supports this diagnosis.
- The family history of **clear cell renal cell carcinoma** in the father and **retinal hemangioblastomas** in the sister, combined with the pheochromocytoma, points to **Von Hippel-Lindau (VHL) disease**. VHL disease is caused by a germline mutation in the **VHL tumor suppressor gene** located on **chromosome 3p25-26**.
*Chromosome 11*
- Mutations on chromosome 11 are associated with **Multiple Endocrine Neoplasia (MEN) type 1**, which includes tumors of the **parathyroid, pituitary, and pancreatic islet cells**, but typically not pheochromocytomas or renal cell carcinoma in this familial pattern.
- While other conditions like **Beckwith-Wiedemann syndrome** and some **leukemias** are linked to chromosome 11, they do not fit the presented clinical picture of pheochromocytoma and VHL-associated cancers.
*Chromosome 2*
- No major familial cancer syndromes or endocrine disorders that would present with the combination of pheochromocytoma, renal cell carcinoma, and retinal hemangioblastomas are primarily linked to chromosome 2.
- While various genetic conditions involve chromosome 2, they do not align with the specific presentation of **VHL disease**.
*Chromosome 17*
- Mutations on chromosome 17 are notably associated with **Neurofibromatosis type 1 (NF1)**, which can present with pheochromocytoma, but typically also involves **café-au-lait spots, neurofibromas, optic gliomas**, and Lisch nodules. The patient's presentation does not describe these characteristic NF1 features.
- **TP53 gene mutations** on chromosome 17 are linked to **Li-Fraumeni syndrome**, predisposing to various cancers, but not typically with the VHL-specific combination described.
*Chromosome 10*
- Mutations on chromosome 10 are associated with **Multiple Endocrine Neoplasia (MEN) type 2**, which includes medullary thyroid cancer and pheochromocytoma, but not renal cell carcinoma or retinal hemangioblastomas.
- The specific array of familial cancers (clear cell renal cell carcinoma, retinal hemangioblastoma) strongly deviates from typical MEN2 presentation.
Chromosomal structure US Medical PG Question 8: A 40-year-old woman brings her 2-day-old infant to the pediatrician’s office for a routine checkup. She tells the pediatrician that her baby vomits a greenish-yellow fluid after every feeding session. She has not been very successful in feeding him due to this problem. She also says that her baby has not passed stool since they left the hospital. On examination, the pediatrician observes that the baby has a flat facial profile and small eyes. The epicanthal folds are prominent and the palms have a single transverse crease. His abdomen is distended with high-pitched bowel sounds. The pediatrician orders an abdominal radiograph, the film is shown in the picture. Which of the following best explains the physical and clinical features exhibited by this infant?
- A. Monosomy
- B. Trisomy (Correct Answer)
- C. Genomic imprinting
- D. Anticipation
- E. Locus heterogeneity
Chromosomal structure Explanation: ***Trisomy***
- The combination of **facial dysmorphism** (flat facial profile, small eyes, prominent epicanthal folds), a **single transverse palmar crease**, and **gastrointestinal obstruction** (vomiting greenish-yellow fluid, abdominal distension, high-pitched bowel sounds, failure to pass stool) strongly points to **Down syndrome (Trisomy 21)**.
- The abdominal radiograph shows a **double-bubble sign**, which is characteristic of **duodenal atresia**, a common congenital anomaly seen in infants with Trisomy 21.
*Monosomy*
- **Monosomy** refers to the absence of one chromosome from a pair. The most common human monosomy compatible with life is **Turner syndrome (Monosomy X)**, which affects females.
- Turner syndrome presents with distinct features like **short stature**, **webbed neck**, and **gonadal dysgenesis**, which are not present in this infant.
*Genomic imprinting*
- **Genomic imprinting** is an epigenetic phenomenon where certain genes are expressed in a **parent-of-origin-specific manner**. Examples include **Prader-Willi** and **Angelman syndromes**.
- These syndromes have specific clinical features (e.g., **hyperphagia** and developmental delay in Prader-Willi, severe intellectual disability and **ataxia** in Angelman) that do not match the infant's presentation.
*Anticipation*
- **Anticipation** is a phenomenon in genetics where the symptoms of a genetic disorder become **more severe** and appear at an **earlier age** in successive generations.
- This typically occurs in disorders caused by the expansion of **trinucleotide repeats**, such as **Huntington's disease** or **myotonic dystrophy**, and does not explain these congenital anomalies.
*Locus heterogeneity*
- **Locus heterogeneity** describes a condition or trait caused by mutations in **different genes** at **different chromosomal loci**.
- While many complex genetic disorders exhibit locus heterogeneity, it primarily explains **inheritance patterns** and does not directly describe the specific clinical and radiographic findings of a single patient with multiple congenital anomalies.
Chromosomal structure US Medical PG Question 9: A 49-year-old man presents to your clinic with “low back pain”. When asked to point to the area that bothers him the most, he motions to both his left and right flank. He describes the pain as deep, dull, and aching for the past few months. His pain does not change significantly with movement or lifting heavy objects. He noted dark colored urine this morning. He has a history of hypertension managed with hydrochlorothiazide; however, he avoids seeing the doctor whenever possible. He drinks 3-4 beers on the weekends but does not smoke. His father died of a sudden onset brain bleed, and his mother has diabetes. In clinic, his temperature is 99°F (37.2°C), blood pressure is 150/110 mmHg, pulse is 95/min, and respirations are 12/min. Bilateral irregular masses are noted on deep palpation of the abdomen. The patient has full range of motion in his back and has no tenderness of the spine or paraspinal muscles. Urine dipstick in clinic is notable for 3+ blood. Which chromosome is most likely affected by a mutation in this patient?
- A. Chromosome 6
- B. Chromosome 7
- C. Chromosome 4
- D. Chromosome 15
- E. Chromosome 16 (Correct Answer)
Chromosomal structure Explanation: ***Chromosome 16***
- This patient's presentation with bilateral flank pain, hypertension, hematuria (dark urine with 3+ blood on dipstick), and palpable bilateral irregular abdominal masses is highly suggestive of **Autosomal Dominant Polycystic Kidney Disease (ADPKD)**.
- The most common form of ADPKD, comprising about 85% of cases, is caused by mutations in the **PKD1 gene** located on **chromosome 16**.
*Chromosome 6*
- Mutations on chromosome 6 are associated with conditions such as **hemochromatosis (HFE gene)** and certain types of **human leukocyte antigen (HLA) linked diseases**, neither of which fits the patient's primary symptoms.
- There is no direct link between chromosome 6 mutations and the classic presentation of ADPKD.
*Chromosome 7*
- Mutations on chromosome 7 are linked to conditions like **Cystic Fibrosis (CFTR gene)** and **Williams-Beuren Syndrome**.
- While CFTR mutations can cause renal cysts in some atypical cases, it does not typically present with the extensive renal manifestations and palpable masses seen in ADPKD.
*Chromosome 4*
- Chromosome 4 harbors the **PKD2 gene**, which is responsible for approximately 15% of ADPKD cases (ADPKD type 2).
- While PKD2 mutations can cause ADPKD, they generally present with a milder phenotype and later onset compared to PKD1 mutations. Given this patient's classic presentation with significant bilateral masses and relatively younger age, PKD1 (chromosome 16) is more likely.
- Chromosome 4 is also associated with **Huntington's disease**.
*Chromosome 15*
- Mutations on chromosome 15 are linked to conditions such as **Marfan syndrome** and **Prader-Willi/Angelman syndromes**.
- These conditions have distinct clinical features that do not align with the patient's symptoms of significant renal pathology.
Chromosomal structure US Medical PG Question 10: A 5-year-old boy is brought to the physician because of behavioral problems. His mother says that he has frequent angry outbursts and gets into fights with his classmates. He constantly complains of feeling hungry, even after eating a full meal. He has no siblings, and both of his parents are healthy. He is at the 25th percentile for height and is above the 95th percentile for weight. Physical examination shows central obesity, undescended testes, almond-shaped eyes, and a thin upper lip. Which of the following genetic changes is most likely associated with this patient's condition?
- A. Mitotic nondisjunction of chromosome 21
- B. Mutation of FBN-1 gene on chromosome 15
- C. Microdeletion of long arm of chromosome 7
- D. Loss of paternal gene expression on chromosome 15 (Correct Answer)
- E. Deletion of Phe508 on chromosome 7
Chromosomal structure Explanation: ***Loss of paternal gene expression on chromosome 15***
- The patient's symptoms, including **hyperphagia**, **obesity**, behavioral issues, short stature, and **hypogonadism** (undescended testes), are characteristic of **Prader-Willi syndrome**.
- Prader-Willi syndrome is most commonly caused by the **loss of paternal gene expression** from the **q11-q13 region of chromosome 15**, either due to a paternal deletion, maternal uniparental disomy, or a defect in the imprinting center.
*Microdeletion of long arm of chromosome 7*
- A microdeletion on the long arm of chromosome 7 (7q11.23) is associated with **Williams syndrome**, characterized by an **elfin facial appearance**, supravalvular aortic stenosis, and intellectual disability.
- This does not match the patient's symptoms of obesity, hyperphagia, or hypogonadism.
*Deletion of Phe508 on chromosome 7*
- A deletion of phenylalanine at position 508 (**ΔF508**) on chromosome 7 is the most common mutation in the **cystic fibrosis transmembrane conductance regulator (CFTR)** gene, causing **cystic fibrosis**.
- Cystic fibrosis is an **autosomal recessive disorder** requiring mutations in both alleles (inherited from both parents), and primarily affects the exocrine glands, leading to lung disease, pancreatic insufficiency, and infertility, which are unrelated to the patient's presentation.
*Mutation of FBN-1 gene on chromosome 15*
- A mutation in the **FBN1 gene** on chromosome 15 (15q21.1) causes **Marfan syndrome**, which is a connective tissue disorder.
- Marfan syndrome presents with tall stature, long limbs (**arachnodactyly**), lens dislocation, and aortic root dilation, none of which are described in this patient.
*Mitotic nondisjunction of chromosome 21*
- Mitotic nondisjunction of chromosome 21 can lead to **mosaic Down syndrome**, but **trisomy 21** (due to meiotic nondisjunction) is the most common cause of Down syndrome.
- Down syndrome is associated with characteristic facial features, intellectual disability, and congenital heart defects, which are distinct from the symptoms presented.
More Chromosomal structure US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.