Homologous recombination US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Homologous recombination. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Homologous recombination US Medical PG Question 1: A 42-year-old woman is seen by her primary care physician for her annual checkup. She has no current concerns and says that she has been healthy over the last year except for a bout of the flu in December. She has no significant past medical history and is not currently taking any medications. She has smoked 1 pack per day since she was 21 and drinks socially with her friends. Her family history is significant for prostate cancer in her dad when he was 51 years of age and ovarian cancer in her paternal aunt when she was 41 years of age. Physical exam reveals a firm, immobile, painless lump in the upper outer quadrant of her left breast as well as 2 smaller nodules in the lower quadrants of her right breast. Biopsy of these lesions shows small, atypical, glandular, duct-like cells with stellate morphology. Which of the following pathways is most likely abnormal in this patient?
- A. Nucleotide excision repair
- B. Base excision repair
- C. Non-homologous end joining
- D. Mismatch repair
- E. Homologous recombination (Correct Answer)
Homologous recombination Explanation: ***Homologous recombination***
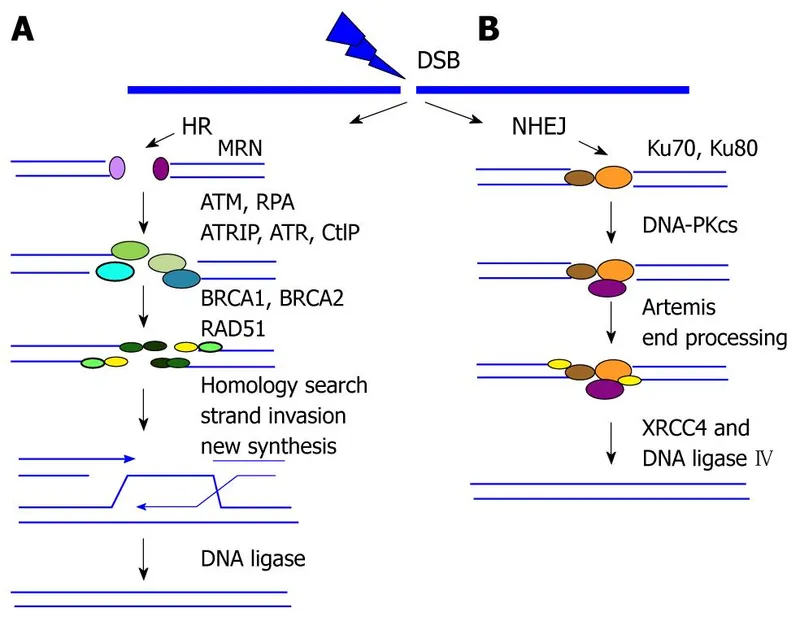
- The patient's presentation with **multiple, bilateral breast lumps** and a strong family history of **early-onset breast, ovarian, and prostate cancers** suggests a hereditary cancer syndrome, most notably related to **BRCA1/2 mutations**.
- **BRCA1 and BRCA2 genes** are crucial for **homologous recombination**, a major pathway for repairing **double-strand DNA breaks**. Defects in this pathway lead to genomic instability and increased cancer risk.
*Nucleotide excision repair*
- This pathway primarily repairs bulky DNA adducts, such as **pyrimidine dimers** caused by UV radiation, and maintains DNA integrity by excising the damaged segment.
- Deficiencies in nucleotide excision repair are associated with diseases like **xeroderma pigmentosum**, which is characterized by extreme sensitivity to sunlight and skin cancers, not the pattern seen in this patient.
*Base excision repair*
- **Base excision repair** is responsible for repairing small, non-bulky DNA lesions, such as **oxidized or deaminated bases**, by removing the damaged base and replacing it.
- While essential for DNA integrity, its malfunction is not typically linked to the **hereditary breast and ovarian cancer syndrome** suggested by the patient's family history and clinical presentation.
*Non-homologous end joining*
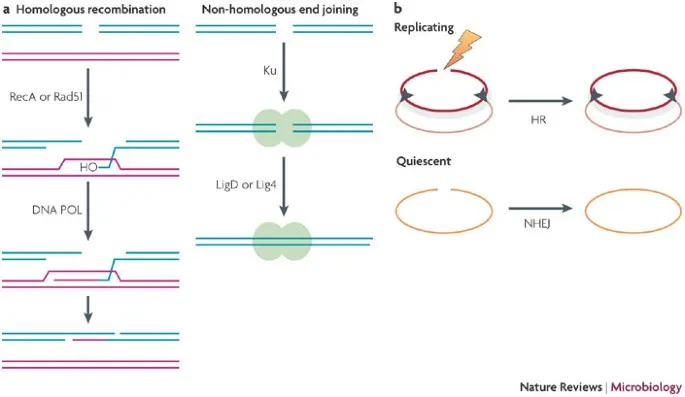
- **Non-homologous end joining (NHEJ)** is an error-prone pathway for repairing **double-strand DNA breaks** by directly ligating the broken ends without a homologous template.
- While critical for DNA repair, defects in NHEJ are not the primary cause of hereditary breast and ovarian cancer, which is more specifically linked to the **BRCA1/2 genes** and the homologous recombination pathway.
*Mismatch repair*
- **Mismatch repair (MMR)** corrects errors that occur during DNA replication, such as mismatched bases or small insertions/deletions.
- Deficiencies in MMR lead to **microsatellite instability** and are characteristic of **Lynch syndrome (hereditary nonpolyposis colorectal cancer)**, which primarily increases the risk of colorectal, endometrial, and other specific cancers, but not the pattern of breast, ovarian, and prostate cancers seen here.
Homologous recombination US Medical PG Question 2: A 3-year-old male child is found to have a disease involving DNA repair. Specifically, he is found to have a defect in the endonucleases involved in the nucleotide excision repair of pyrimidine dimers. Which of the following is a unique late-stage complication of this child's disease?
- A. Telangiectasia
- B. Colorectal cancer
- C. Malignant melanoma (Correct Answer)
- D. Lymphomas
- E. Endometrial cancer
Homologous recombination Explanation: **Malignant melanoma**
- The described condition is **xeroderma pigmentosum**, an autosomal recessive disorder characterized by a defect in **nucleotide excision repair (NER)**, specifically the inability to remove **pyrimidine dimers** caused by **UV radiation**.
- This severely impaired DNA repair leads to an extreme predisposition to **UV-induced skin cancers**, including basal cell carcinomas, squamous cell carcinomas, and, most aggressively, **malignant melanoma**, which is a unique and life-threatening late-stage complication.
*Telangiectasia*
- **Telangiectasias** are dilated small blood vessels that appear on the skin or mucous membranes and can be associated with various conditions.
- While skin abnormalities are prevalent in xeroderma pigmentosum due to sun damage, **melanoma** is a more specific and severe late-stage complication directly resulting from the DNA repair defect.
*Colorectal cancer*
- **Colorectal cancer** is typically associated with other DNA repair defects, such as those in the **mismatch repair system**, as seen in conditions like **Lynch syndrome**.
- It is not a primary or most significant late-stage complication of xeroderma pigmentosum, which is primarily characterized by skin cancers.
*Lymphomas*
- **Lymphomas** are cancers of the lymphatic system, often linked to immune deficiencies or specific genetic translocations.
- While individuals with genetic syndromes can have increased cancer risks, **lymphoma** is not the hallmark late-stage complication of xeroderma pigmentosum; skin cancers are the predominant concern.
*Endometrial cancer*
- **Endometrial cancer** is a gynecological cancer often associated with hormonal factors or genetic predispositions like Lynch syndrome, which involves mismatch repair defects.
- This type of cancer is not a characteristic or unique late-stage complication of xeroderma pigmentosum, whose pathology is centered on **UV-induced DNA damage** and subsequent skin malignancies.
Homologous recombination US Medical PG Question 3: While performing a Western blot, a graduate student spilled a small amount of the radiolabeled antibody on her left forearm. Although very little harm was done to the skin, the radiation did cause minor damage to the DNA of the exposed skin by severing covalent bonds between the nitrogenous bases and the deoxyribose sugar, leaving several apurinic/apyrimidinic sites. Damaged cells would most likely repair these sites by which of the following mechanisms?
- A. Nucleotide excision repair
- B. Nonhomologous end joining repair
- C. Homologous recombination
- D. Mismatch repair
- E. Base excision repair (Correct Answer)
Homologous recombination Explanation: **Base excision repair**
- This mechanism is specifically involved in correcting **single-base DNA damage** or **modified bases**, such as **apurinic/apyrimidinic (AP) sites**.
- It involves removing the damaged base by a **DNA glycosylase**, creating an AP site, which is then processed by an **AP endonuclease** to cleave the phosphodiester backbone, followed by DNA polymerase and ligase.
*Nucleotide excision repair*
- Primarily repairs **bulky DNA lesions**, such as **thymine dimers** caused by UV radiation, or damage from chemical adducts that distort the DNA helix.
- It involves excising a larger oligonucleotide containing the damage, not just a single base.
*Nonhomologous end joining repair*
- This pathway is used to repair **double-strand DNA breaks**, where both strands of the DNA molecule are broken.
- It is a "quick-and-dirty" repair mechanism that ligates the broken ends together, often leading to small insertions or deletions.
*Homologous recombination*
- A repair mechanism for **double-strand DNA breaks** that uses a homologous DNA template (e.g., sister chromatid) to accurately repair the break.
- This process is highly accurate but occurs only when a homologous template is available, typically during the S and G2 phases of the cell cycle.
*Mismatch repair*
- Corrects **base-pair mismatches** and **small insertions/deletions** that occur during DNA replication, which were not corrected by DNA polymerase proofreading.
- It targets newly synthesized DNA strands based on methylation patterns in the parental strand.
Homologous recombination US Medical PG Question 4: DNA replication is a highly complex process where replication occurs on both strands of DNA. On the leading strand of DNA, replication occurs uninterrupted, but on the lagging strand, replication is interrupted and occurs in fragments called Okazaki fragments. These fragments need to be joined, which of the following enzymes is involved in the penultimate step before ligation can occur?
- A. DNA gyrase
- B. DNA ligase
- C. DNA helicase
- D. DNA polymerase I (Correct Answer)
- E. DNA polymerase III
Homologous recombination Explanation: **DNA polymerase I**
- **DNA polymerase I** plays a crucial role in removing the **RNA primers** from the Okazaki fragments on the lagging strand.
- After primer removal, it fills the resulting gaps with **deoxyribonucleotides** before DNA ligase seals the nicks.
*DNA gyrase*
- **DNA gyrase** (a type of **topoisomerase**) is involved in relieving **supercoiling** ahead of the replication fork.
- It does not directly participate in the joining of Okazaki fragments, but rather in maintaining DNA topology during replication.
*DNA ligase*
- **DNA ligase** is responsible for the **final sealing** of the nicks between adjacent Okazaki fragments.
- It forms a **phosphodiester bond** between the 3'-hydroxyl end of one fragment and the 5'-phosphate end of the next, following primer removal and gap filling.
*DNA helicase*
- **DNA helicase** unwinds the double-stranded DNA helix, separating the two strands at the **replication fork**.
- This enzyme is essential for initiating replication but does not participate in processing Okazaki fragments.
*DNA polymerase III*
- **DNA polymerase III** is the primary enzyme responsible for the **elongation of new DNA strands** in both leading and lagging strand synthesis.
- It synthesizes the actual Okazaki fragments but does not directly remove primers or fill the gaps.
Homologous recombination US Medical PG Question 5: A 34-year-old woman comes to the physician for evaluation of a breast lump she noticed 2 days ago while showering. She has no history of major illness. Her mother died of ovarian cancer at age 38, and her sister was diagnosed with breast cancer at age 33. Examination shows a 1.5-cm, nontender, mobile mass in the upper outer quadrant of the left breast. Mammography shows pleomorphic calcifications. Biopsy of the mass shows invasive ductal carcinoma. The underlying cause of this patient's condition is most likely a mutation of a gene involved in which of the following cellular events?
- A. Repair of double-stranded DNA breaks (Correct Answer)
- B. Inhibition of programmed cell death
- C. Regulation of intercellular adhesion
- D. Activity of cytoplasmic tyrosine kinase
- E. Arrest of cell cycle in G1 phase
Homologous recombination Explanation: ***Repair of double-stranded DNA breaks***
- The patient's **family history** (mother with ovarian cancer at 38, sister with breast cancer at 33) and early onset of **invasive ductal carcinoma** strongly suggest an inherited cancer syndrome.
- **BRCA1 and BRCA2 genes** are tumor suppressor genes responsible for repairing **double-stranded DNA breaks**, and mutations in these genes significantly increase the risk of breast and ovarian cancers.
*Inhibition of programmed cell death*
- Mutations leading to the **inhibition of programmed cell death (apoptosis)**, such as those affecting the **Bcl-2 gene**, can contribute to cancer by allowing damaged cells to survive and proliferate.
- While relevant to cancer pathogenesis, it is not the primary mechanism associated with the specific familial breast/ovarian cancer pattern seen here, which points more directly to DNA repair defects.
*Regulation of intercellular adhesion*
- Defects in **intercellular adhesion**, often involving **E-cadherin** (CDH1 gene) mutations, are associated with cancers like **lobular breast carcinoma** and **hereditary diffuse gastric cancer**.
- This patient has **invasive ductal carcinoma**, and the specific familial pattern is less characteristic of intercellular adhesion defects.
*Activity of cytoplasmic tyrosine kinase*
- Abnormal **cytoplasmic tyrosine kinase activity** is implicated in various cancers (e.g., **HER2/neu** amplification in breast cancer, **BCR-ABL** fusion in CML).
- While HER2/neu overexpression is common in breast cancer, it is typically a somatic mutation or amplification, and not the underlying germline defect explaining the strong family history of early-onset breast and ovarian cancer.
*Arrest of cell cycle in G1 phase*
- The **arrest of the cell cycle at the G1 phase** is mainly regulated by **p53** and **Rb tumor suppressor genes**, which prevent uncontrolled cell division.
- While mutations in these genes are crucial in many cancers, the specific familial pattern (breast and ovarian cancer) points more strongly to defects in homologous recombination via BRCA1/2, a different DNA repair pathway.
Homologous recombination US Medical PG Question 6: A 54-year-old woman with breast cancer comes to the physician because of redness and pain in the right breast. She has been undergoing ionizing radiation therapy daily for the past 2 weeks as adjuvant treatment for her breast cancer. Physical examination shows erythema, edema, and superficial desquamation of the skin along the right breast at the site of radiation. Sensation to light touch is intact. Which of the following is the primary mechanism of DNA repair responsible for preventing radiation-induced damage to neighboring neurons?
- A. Homology-directed repair
- B. Base excision repair
- C. Nonhomologous end joining repair (Correct Answer)
- D. DNA mismatch repair
- E. Nucleotide excision repair
Homologous recombination Explanation: ***Nonhomologous end joining repair***
- This pathway is crucial for repairing **double-strand DNA breaks**, which are a major form of damage caused by **ionizing radiation**.
- It directly ligates the broken DNA ends without requiring a homologous template, making it an efficient but potentially error-prone repair mechanism.
*Homology-directed repair*
- This pathway is also used to repair **double-strand DNA breaks** but requires a **homologous DNA template** (usually a sister chromatid) for accurate repair.
- While highly accurate, it is typically active during the S and G2 phases of the cell cycle and is generally slower and less dominant than NHEJ for immediate radiation-induced damage in non-dividing cells like neurons.
*Base excision repair*
- This mechanism primarily corrects damage to individual DNA bases, such as **oxidative damage**, alkylation, or deamination.
- It is not the primary mechanism for repairing the **double-strand breaks** induced by ionizing radiation.
*DNA mismatch repair*
- This pathway corrects errors that arise during **DNA replication**, specifically mismatched base pairs or small insertions/deletions.
- It is not involved in repairing radiation-induced DNA damage like **double-strand breaks**.
*Nucleotide excision repair*
- This pathway repairs bulky DNA lesions, such as those caused by **UV radiation** (e.g., pyrimidine dimers) or chemical mutagens.
- It removes a segment of DNA containing the damage but is not the primary repair mechanism for **double-strand breaks** caused by ionizing radiation.
Homologous recombination US Medical PG Question 7: An 11-year-old boy is brought to the physician for the evaluation of frequent falling. His mother reports that the patient has had increased difficulty walking over the last few months and has refused to eat solid foods for the past 2 weeks. He has met all developmental milestones. The patient has had multiple ear infections since birth. His temperature is 37°C (98.6°F), pulse is 90/min, and blood pressure is 120/80 mm Hg. Examination shows foot inversion with hammertoes bilaterally. His gait is wide-based with irregular and uneven steps. Laboratory studies show a serum glucose concentration of 300 mg/dL. Further evaluation of this patient is most likely to show which of the following findings?
- A. Expansion of GAA trinucleotide repeats (Correct Answer)
- B. Duplication of PMP22 gene
- C. Mutation of type I collagen gene
- D. Absence of dystrophin protein
- E. Defect of ATM protein
Homologous recombination Explanation: ***Expansion of GAA trinucleotide repeats***
- The clinical presentation of **Friedreich ataxia** includes progressive **ataxia**, **dysarthria**, **hypertrophic cardiomyopathy**, and **diabetes mellitus** (indicated by serum glucose of 300 mg/dL), consistent with the patient's symptoms.
- This condition is caused by an autosomal recessive **GAA trinucleotide repeat expansion** in the frataxin gene.
*Duplication of PMP22 gene*
- A duplication of the **PMP22 gene** is associated with **Charcot-Marie-Tooth disease type 1A**, which presents with progressive distal weakness, sensory loss, foot deformities (e.g., hammertoes, pes cavus), and a steppage gait.
- While some features overlap, the severe **ataxia** and **diabetes mellitus** seen in this patient are not characteristic of Charcot-Marie-Tooth disease.
*Mutation of type I collagen gene*
- **Type I collagen gene mutations** are characteristic of **osteogenesis imperfecta**, a connective tissue disorder causing brittle bones, blue sclerae, and hearing loss.
- The patient's symptoms of neurological decline and diabetes are not consistent with osteogenesis imperfecta.
*Absence of dystrophin protein*
- An absence of **dystrophin protein** is the hallmark of **Duchenne muscular dystrophy**, an X-linked recessive disorder causing progressive muscle weakness, Gowers' sign, and pseudohypertrophy of the calves.
- While muscle weakness would cause falling, the patient's presentation with **ataxia** and a **wide-based gait** points away from a primary muscle disorder.
*Defect of ATM protein*
- A **defect in ATM protein** is responsible for **Ataxia-telangiectasia**, an autosomal recessive disorder characterized by cerebellar ataxia, telangiectasias, immunodeficiency, and an increased risk of cancer.
- While ataxia is present, the patient's findings of **hammertoes** and **diabetes mellitus** are not typical features of Ataxia-telangiectasia.
Homologous recombination US Medical PG Question 8: A software engineer presents to the OPD with 'complaints of easy fatigability. He reports sitting in front of a computer for 12-14 hours a day consuming junk food, and eating few fruits and vegetables. CBC results show hemoglobin (Hb) concentration of $7 \mathrm{gm} \%$ and MCV of 120 fL . What is the most likely cause of anemia?
- A. Cyanocobalamin deficiency
- B. Acute blood loss
- C. Sideroblastic anemia
- D. Folate deficiency (Correct Answer)
- E. Iron deficiency anemia
Homologous recombination Explanation: ***Folate deficiency***
- A **macrocytic anemia** with an **MCV of 120 fL** is characteristic of folate deficiency, as folate is vital for **DNA synthesis** in red blood cell production.
- The patient's diet of **junk food** and few fruits/vegetables suggests poor nutritional intake, as folate is abundant in leafy greens and fresh produce.
*Cyanocobalamin deficiency*
- While also causing **macrocytic anemia** with high MCV, cyanocobalamin (Vitamin B12) deficiency often presents with **neurological symptoms** (e.g., neuropathy, cognitive changes) which are not mentioned.
- Dietary sources of B12 are primarily **animal products**, and while junk food is poor, a strict vegetarian/vegan diet is a stronger indicator of B12 deficiency.
*Acute blood loss*
- Acute blood loss typically causes **normocytic, normochromic anemia**, characterized by a normal MCV in the initial stages.
- While severe blood loss can lead to fatigue, the **elevated MCV** of 120 fL makes this diagnosis unlikely unless there's a pre-existing macrocytic condition.
*Sideroblastic anemia*
- Sideroblastic anemia can be **microcytic, normocytic, or macrocytic**, but it is primarily characterized by the presence of **ring sideroblasts** in the bone marrow and iron overload.
- It's often associated with **alcoholism, lead poisoning, or myelodysplastic syndromes**, and the typical features of the patient's diet and MCV do not point towards this condition.
*Iron deficiency anemia*
- Iron deficiency anemia presents with **microcytic, hypochromic anemia** with a **low MCV** (typically <80 fL), not macrocytic anemia.
- While iron deficiency is the most common cause of anemia worldwide and can result from poor diet, the **elevated MCV of 120 fL** clearly excludes this diagnosis.
Homologous recombination US Medical PG Question 9: An investigator isolates bacteria from a patient who presented with dysuria and urinary frequency. These bacteria grow rapidly in pink colonies on MacConkey agar. During replication of these bacteria, the DNA strands are unwound at the origin of replication, forming two Y-shaped replication forks that open in opposite directions. At each replication fork, daughter strands are synthesized from the template strands in a 5′ to 3′ direction. On one strand, the DNA is synthesized continuously; on the other strand, the DNA is synthesized in short segments. The investigator finds that three enzymes are directly involved in elongating the DNA of the lagging strand in these bacteria. One of these enzymes has an additional function that the others do not possess. Which of the following steps in DNA replication is unique to this enzyme?
- A. Elongation of lagging strand in 5'→3' direction
- B. Excision of nucleotides with 5'→3' exonuclease activity (Correct Answer)
- C. Prevention of reannealing of the leading strand and the lagging strand
- D. Creation of ribonucleotide primers
- E. Proofreading for mismatched nucleotides
Homologous recombination Explanation: ***Excision of nucleotides with 5'→3' exonuclease activity***
- **DNA polymerase I** possesses unique **5'→3' exonuclease activity** that allows it to remove RNA primers synthesized by primase.
- After primer removal, DNA polymerase I synthesizes DNA in the 5'→3' direction to fill the gap.
*Elongation of lagging strand in 5'→3' direction*
- While **DNA polymerase I** elongates the lagging strand, this 5'→3' synthesis function is also shared by **DNA polymerase III**, which is the primary enzyme for DNA synthesis.
- Therefore, this specific function is not unique to the enzyme in question (DNA polymerase I) as DNA polymerase III also performs 5'→3' elongation.
*Prevention of reannealing of the leading strand and the lagging strand*
- This function is carried out by **single-strand binding proteins (SSBs)**, which bind to the separated DNA strands to prevent them from reannealing and protect them from degradation.
- This is not a function of any DNA polymerase.
*Creation of ribonucleotide primers*
- The synthesis of short RNA primers required for initiation of DNA synthesis is performed by **primase**, an RNA polymerase.
- DNA polymerases do not create primers but rather extend them.
*Proofreading for mismatched nucleotides*
- **DNA polymerase I** and **DNA polymerase III** both possess **3'→5' exonuclease activity** for proofreading, allowing them to remove incorrectly incorporated nucleotides.
- Since this function is shared by DNA polymerase III, it is not unique to DNA polymerase I.
Homologous recombination US Medical PG Question 10: An investigator is studying the replication of bacterial DNA with modified nucleotides. After unwinding, the double-stranded DNA forms a Y-shaped replication fork that separates into two strands. At each of these strands, daughter strands are synthesized. One strand is continuously extended from the template strands in a 5′ to 3′ direction. Which of the following is exclusively associated with the strand being synthesized away from the replication fork?
- A. Reverse transcriptase activity
- B. Repeated activity of ligase (Correct Answer)
- C. Elongation in the 3'→5' direction
- D. Synthesis of short RNA sequences
- E. 5' → 3' exonuclease activity
Homologous recombination Explanation: ***Repeated activity of ligase***
- The lagging strand, synthesized away from the replication fork, is made in fragments (**Okazaki fragments**) due to the 5' to 3' synthesis direction of DNA polymerase.
- **DNA ligase** repeatedly joins these Okazaki fragments together, forming a continuous strand.
*Reverse transcriptase activity*
- **Reverse transcriptase** synthesizes DNA from an RNA template, which is not involved in normal bacterial DNA replication.
- This enzyme is characteristic of **retroviruses** and certain eukaryotic telomere maintenance.
*Elongation in the 3'→5' direction*
- DNA polymerases only synthesize new DNA strands in the **5' to 3' direction**.
- While reading the template strand in the 3' to 5' direction, the daughter strand is always built from 5' to 3'.
*Synthesis of short RNA sequences*
- **RNA primers** are synthesized by **primase** on both the leading and lagging strands to initiate DNA synthesis.
- This process is not exclusive to the strand synthesized away from the replication fork; the leading strand also requires an initial RNA primer.
*5' → 3' exonuclease activity*
- The **5' to 3' exonuclease activity** of DNA polymerase I in bacteria is responsible for removing RNA primers.
- This activity occurs on both the leading and lagging strands, as both require primer removal.
More Homologous recombination US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.