Base excision repair US Medical PG Practice Questions and MCQs
Practice US Medical PG questions for Base excision repair. These multiple choice questions (MCQs) cover important concepts and help you prepare for your exams.
Base excision repair US Medical PG Question 1: A 3-year-old male child is found to have a disease involving DNA repair. Specifically, he is found to have a defect in the endonucleases involved in the nucleotide excision repair of pyrimidine dimers. Which of the following is a unique late-stage complication of this child's disease?
- A. Telangiectasia
- B. Colorectal cancer
- C. Malignant melanoma (Correct Answer)
- D. Lymphomas
- E. Endometrial cancer
Base excision repair Explanation: **Malignant melanoma**
- The described condition is **xeroderma pigmentosum**, an autosomal recessive disorder characterized by a defect in **nucleotide excision repair (NER)**, specifically the inability to remove **pyrimidine dimers** caused by **UV radiation**.
- This severely impaired DNA repair leads to an extreme predisposition to **UV-induced skin cancers**, including basal cell carcinomas, squamous cell carcinomas, and, most aggressively, **malignant melanoma**, which is a unique and life-threatening late-stage complication.
*Telangiectasia*
- **Telangiectasias** are dilated small blood vessels that appear on the skin or mucous membranes and can be associated with various conditions.
- While skin abnormalities are prevalent in xeroderma pigmentosum due to sun damage, **melanoma** is a more specific and severe late-stage complication directly resulting from the DNA repair defect.
*Colorectal cancer*
- **Colorectal cancer** is typically associated with other DNA repair defects, such as those in the **mismatch repair system**, as seen in conditions like **Lynch syndrome**.
- It is not a primary or most significant late-stage complication of xeroderma pigmentosum, which is primarily characterized by skin cancers.
*Lymphomas*
- **Lymphomas** are cancers of the lymphatic system, often linked to immune deficiencies or specific genetic translocations.
- While individuals with genetic syndromes can have increased cancer risks, **lymphoma** is not the hallmark late-stage complication of xeroderma pigmentosum; skin cancers are the predominant concern.
*Endometrial cancer*
- **Endometrial cancer** is a gynecological cancer often associated with hormonal factors or genetic predispositions like Lynch syndrome, which involves mismatch repair defects.
- This type of cancer is not a characteristic or unique late-stage complication of xeroderma pigmentosum, whose pathology is centered on **UV-induced DNA damage** and subsequent skin malignancies.
Base excision repair US Medical PG Question 2: A 62-year-old man comes to the physician because of progressive fatigue and dyspnea on exertion for 3 months. During this time, he has also had increased straining during defecation and a 10-kg (22-lb) weight loss. He has no personal or family history of serious medical illness. Physical examination shows conjunctival pallor. Laboratory studies show microcytic anemia. Test of the stool for occult blood is positive. Colonoscopy shows an exophytic mass in the ascending colon. Pathologic examination of the mass shows a well-differentiated adenocarcinoma. A gain-of-function mutation in which of the following genes is most likely involved in the pathogenesis of this patient's condition?
- A. APC
- B. TP53
- C. MLH1
- D. KRAS (Correct Answer)
- E. DCC
Base excision repair Explanation: ***KRAS***
- A **gain-of-function mutation** in **KRAS** is a common early event in the development of colorectal adenocarcinoma, driving uncontrolled cell growth and proliferation.
- This mutation is frequently found in **sporadic colorectal cancers**, particularly in the advanced stages of adenoma to carcinoma progression.
*APC*
- **APC** is a **tumor suppressor gene**, and mutations in it are typically **loss-of-function**, not gain-of-function.
- While APC mutations are crucial early steps in the adenoma-carcinoma sequence, they lead to inactivation of the gene, not increased function.
*TP53*
- **TP53** is a **tumor suppressor gene** which, when mutated, usually involves **loss-of-function** or dominant-negative effects, impairing its ability to induce apoptosis or cell cycle arrest.
- Mutations in TP53 are typically associated with **later stages** of colorectal cancer progression and tend to be loss-of-function, not gain-of-function.
*MLH1*
- **MLH1** is involved in **DNA mismatch repair**, and mutations here lead to **microsatellite instability** and are characteristic of hereditary nonpolyposis colorectal cancer (Lynch syndrome).
- These are typically **loss-of-function mutations** that impair DNA repair, not gain-of-function mutations promoting oncogenesis directly through signaling pathways.
*DCC*
- **DCC** (**Deleted in Colorectal Carcinoma**) is a **tumor suppressor gene**, and its inactivation or loss is associated with colorectal cancer progression, particularly the transition from adenoma to carcinoma.
- Mutations or deletions in DCC result in a **loss-of-function**, not a gain-of-function, contributing to tumor growth by failing to regulate cell differentiation and apoptosis.
Base excision repair US Medical PG Question 3: While performing a Western blot, a graduate student spilled a small amount of the radiolabeled antibody on her left forearm. Although very little harm was done to the skin, the radiation did cause minor damage to the DNA of the exposed skin by severing covalent bonds between the nitrogenous bases and the deoxyribose sugar, leaving several apurinic/apyrimidinic sites. Damaged cells would most likely repair these sites by which of the following mechanisms?
- A. Nucleotide excision repair
- B. Nonhomologous end joining repair
- C. Homologous recombination
- D. Mismatch repair
- E. Base excision repair (Correct Answer)
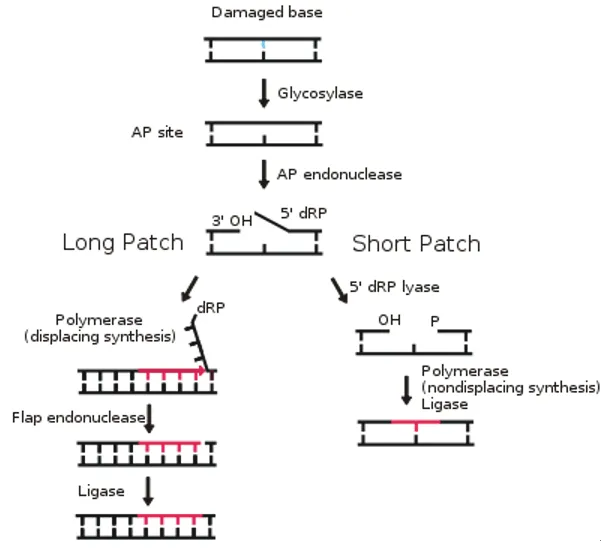
Base excision repair Explanation: **Base excision repair**
- This mechanism is specifically involved in correcting **single-base DNA damage** or **modified bases**, such as **apurinic/apyrimidinic (AP) sites**.
- It involves removing the damaged base by a **DNA glycosylase**, creating an AP site, which is then processed by an **AP endonuclease** to cleave the phosphodiester backbone, followed by DNA polymerase and ligase.
*Nucleotide excision repair*
- Primarily repairs **bulky DNA lesions**, such as **thymine dimers** caused by UV radiation, or damage from chemical adducts that distort the DNA helix.
- It involves excising a larger oligonucleotide containing the damage, not just a single base.
*Nonhomologous end joining repair*
- This pathway is used to repair **double-strand DNA breaks**, where both strands of the DNA molecule are broken.
- It is a "quick-and-dirty" repair mechanism that ligates the broken ends together, often leading to small insertions or deletions.
*Homologous recombination*
- A repair mechanism for **double-strand DNA breaks** that uses a homologous DNA template (e.g., sister chromatid) to accurately repair the break.
- This process is highly accurate but occurs only when a homologous template is available, typically during the S and G2 phases of the cell cycle.
*Mismatch repair*
- Corrects **base-pair mismatches** and **small insertions/deletions** that occur during DNA replication, which were not corrected by DNA polymerase proofreading.
- It targets newly synthesized DNA strands based on methylation patterns in the parental strand.
Base excision repair US Medical PG Question 4: A 14-year-old boy presents to his pediatrician with a 5-day history of abdominal pain and bloody stool. He denies having a fever and says that he has not experienced any other symptoms associated with the abdominal pain. He has no past medical history and does not take any medications or supplements. His family history is significant for a grandfather who developed Alzheimer disease at age 80 and a cousin who died at age 21 from colon cancer. Physical exam is unremarkable. Based on clinical suspicion a colonoscopy is obtained showing hundreds of small polyps in the colon. A mutation of a gene on which of the following chromosomes is most likely responsible for this patient's symptoms?
- A. 5 (Correct Answer)
- B. 7
- C. X
- D. 17
- E. 19
Base excision repair Explanation: ***Correct: Chromosome 5***
- The patient's presentation with hundreds of small colon polyps on colonoscopy strongly suggests **Familial Adenomatous Polyposis (FAP)**
- FAP is an **autosomal dominant** disorder caused by a mutation in the **adenomatous polyposis coli (APC) gene**, which is located on **chromosome 5 (5q21-q22)**
- The family history of a cousin dying at age 21 from colon cancer supports this diagnosis
*Incorrect: Chromosome 7*
- Mutations on chromosome 7 are not typically associated with FAP or extensive colonic polyposis
- While other genetic conditions are linked to chromosome 7 (e.g., cystic fibrosis), they do not present with this specific phenotype
*Incorrect: Chromosome X*
- Mutations on the X chromosome cause **X-linked disorders**, affecting males disproportionately
- Hereditary colorectal cancer syndromes are generally autosomal, not X-linked
*Incorrect: Chromosome 17*
- Chromosome 17 is associated with several genetic conditions, including **Neurofibromatosis type 1 (NF1)** and mutations in **TP53 (Li-Fraumeni syndrome)**
- While Li-Fraumeni syndrome increases cancer risk, it typically presents with a broader spectrum of cancers and not primarily with hundreds of colonic polyps in adolescence
*Incorrect: Chromosome 19*
- Chromosome 19 contains genes linked to various conditions, but not typically to FAP or other hereditary polyposis syndromes of the colon
- For instance, **myotonic dystrophy type 1** is linked to chromosome 19, which does not involve extensive colonic polyps
Base excision repair US Medical PG Question 5: A 38-year-old man presents to his primary care practitioner for 2 months of rectal bleeding. He also reports occasional diarrhea and abdominal pain. His family history is relevant for his father and uncle, who died from complications of colorectal cancer. Colonoscopy shows more than 10 colorectal adenomas. Which of the following genes is most likely affected in this patient?
- A. RAS
- B. TP53
- C. hMLH1
- D. PPAR
- E. APC (Correct Answer)
Base excision repair Explanation: ***APC***
- This patient's presentation with **numerous colorectal adenomas** (over 10), early-onset symptoms (38 years old), and a strong **family history of colorectal cancer** (father and uncle) is highly characteristic of **Familial Adenomatous Polyposis (FAP)**.
- FAP is an **autosomal dominant** condition caused by a germline mutation in the **APC tumor suppressor gene**, leading to the development of hundreds to thousands of adenomatous polyps in the colon, which inevitably progress to colorectal cancer if untreated.
*RAS*
- **RAS mutations** are commonly found in sporadic colorectal cancers and play a role in tumor growth and progression, but they are not typically associated with the **hereditary syndrome of multiple adenomas** seen in this patient.
- RAS activation leads to an increase in **cell proliferation** and can contribute to the development of many cancers, but not as the primary genetic defect in a polyposis syndrome.
*TP53*
- **TP53** is a well-known tumor suppressor gene, and mutations are involved in various cancers, including colorectal cancer (often in its later stages). However, germline mutations in TP53 are associated with **Li-Fraumeni syndrome**, which involves a broad spectrum of early-onset cancers and is not primarily characterized by numerous colonic adenomas.
- TP53 mutations are generally hallmarks of **genomic instability** and are more often seen in the progression of sporadic cancers rather than initiating a polyposis syndrome.
*hMLH1*
- **hMLH1** is a gene involved in **DNA mismatch repair**. Germline mutations in this gene, along with other mismatch repair genes (e.g., MSH2, MSH6, PMS2), are responsible for **Lynch syndrome (hereditary non-polyposis colorectal cancer - HNPCC)**.
- While Lynch syndrome is an important cause of hereditary colorectal cancer, it is characterized by fewer polyps (typically <10) that progress rapidly to cancer, and an increased risk of other cancers (e.g., endometrial), which differs from the presentation of **hundreds of adenomas** seen in FAP.
*PPAR*
- **PPARs (Peroxisome Proliferator-Activated Receptors)** are a group of nuclear receptor proteins that play roles in metabolism, cell differentiation, and inflammation.
- While PPAR pathways have been investigated for their potential role in cancer development and as therapeutic targets, **mutations in PPAR genes are not directly linked** to a common hereditary colorectal cancer syndrome characterized by numerous adenomas like FAP.
Base excision repair US Medical PG Question 6: A 54-year-old woman with breast cancer comes to the physician because of redness and pain in the right breast. She has been undergoing ionizing radiation therapy daily for the past 2 weeks as adjuvant treatment for her breast cancer. Physical examination shows erythema, edema, and superficial desquamation of the skin along the right breast at the site of radiation. Sensation to light touch is intact. Which of the following is the primary mechanism of DNA repair responsible for preventing radiation-induced damage to neighboring neurons?
- A. Homology-directed repair
- B. Base excision repair
- C. Nonhomologous end joining repair (Correct Answer)
- D. DNA mismatch repair
- E. Nucleotide excision repair
Base excision repair Explanation: ***Nonhomologous end joining repair***
- This pathway is crucial for repairing **double-strand DNA breaks**, which are a major form of damage caused by **ionizing radiation**.
- It directly ligates the broken DNA ends without requiring a homologous template, making it an efficient but potentially error-prone repair mechanism.
*Homology-directed repair*
- This pathway is also used to repair **double-strand DNA breaks** but requires a **homologous DNA template** (usually a sister chromatid) for accurate repair.
- While highly accurate, it is typically active during the S and G2 phases of the cell cycle and is generally slower and less dominant than NHEJ for immediate radiation-induced damage in non-dividing cells like neurons.
*Base excision repair*
- This mechanism primarily corrects damage to individual DNA bases, such as **oxidative damage**, alkylation, or deamination.
- It is not the primary mechanism for repairing the **double-strand breaks** induced by ionizing radiation.
*DNA mismatch repair*
- This pathway corrects errors that arise during **DNA replication**, specifically mismatched base pairs or small insertions/deletions.
- It is not involved in repairing radiation-induced DNA damage like **double-strand breaks**.
*Nucleotide excision repair*
- This pathway repairs bulky DNA lesions, such as those caused by **UV radiation** (e.g., pyrimidine dimers) or chemical mutagens.
- It removes a segment of DNA containing the damage but is not the primary repair mechanism for **double-strand breaks** caused by ionizing radiation.
Base excision repair US Medical PG Question 7: A group of microbiological investigators is studying bacterial DNA replication in E. coli colonies. While the cells are actively proliferating, the investigators stop the bacterial cell cycle during S phase and isolate an enzyme involved in DNA replication. An assay of the enzyme's exonuclease activity determines that it is active on both intact and demethylated thymine nucleotides. Which of the following enzymes have the investigators most likely isolated?
- A. DNA ligase
- B. Telomerase
- C. Primase
- D. DNA topoisomerase
- E. DNA polymerase I (Correct Answer)
Base excision repair Explanation: ***DNA polymerase I***
- **DNA polymerase I** possesses **5' to 3' exonuclease activity**, which is crucial for removing **RNA primers** (intact nucleotides) laid down by primase during DNA replication.
- This 5' to 3' exonuclease activity also allows it to excise damaged DNA, including DNA containing **demethylated thymine nucleotides**.
- It also has 3' to 5' exonuclease activity for proofreading.
- **Key distinction:** While DNA polymerase III (the main replicative enzyme) only has 3' to 5' exonuclease activity, DNA polymerase I has **both** 3' to 5' and 5' to 3' exonuclease activities, making it essential for primer removal and DNA repair.
*DNA ligase*
- **DNA ligase** functions to form a **phosphodiester bond** between adjacent nucleotides to seal nicks in the DNA backbone, but it does not have exonuclease activity.
- Its primary role is in joining Okazaki fragments and repairing single-strand breaks.
*Telomerase*
- **Telomerase** is a specialized reverse transcriptase that extends the telomeres at the ends of eukaryotic chromosomes, but is not present in prokaryotes like *E. coli*.
- It uses an RNA template to synthesize DNA, and it lacks exonuclease activity.
*Primase*
- **Primase** is an RNA polymerase that synthesizes short **RNA primers** on the DNA template, providing a starting point for DNA synthesis.
- It is involved in synthesizing primers, not in removing or excising nucleotides, and has no exonuclease activity.
*DNA topoisomerase*
- **DNA topoisomerases** relieve supercoiling in DNA during replication and transcription by cutting and rejoining DNA strands.
- While they act on DNA, their function is to manage topological stress, and they do not exhibit exonuclease activity on nucleotides.
Base excision repair US Medical PG Question 8: A 47-year-old man presents to his primary care physician for fatigue. Over the past 3 months, his tiredness has impacted his ability to work as a corporate lawyer. He denies any changes to his diet, exercise regimen, bowel movements, or urinary frequency. His past medical history is notable for obesity, type II diabetes mellitus, and hypertension. He takes metformin and enalapril. His family history is notable for colorectal cancer in his father and paternal grandfather and endometrial cancer in his paternal aunt. He has a 20-pack-year smoking history and drinks one 6-pack of beer a week. His temperature is 98.8°F (37.1°C), blood pressure is 129/71 mmHg, pulse is 82/min, and respirations are 17/min. On exam, he has conjunctival pallor. A stool sample is positive for occult blood. A colonoscopy reveals a small hemorrhagic mass at the junction of the ascending and transverse colon. Which of the following processes is likely impaired in this patient?
- A. Mismatch repair (Correct Answer)
- B. Homologous recombination
- C. Non-homologous end joining
- D. Nucleotide excision repair
- E. Base excision repair
Base excision repair Explanation: ***Mismatch repair***
- The patient's presentation with **colorectal cancer** at a relatively young age and a strong family history of various cancers (colorectal, endometrial) in **first-degree and second-degree relatives** suggests Lynch syndrome (Hereditary Nonpolyposis Colorectal Cancer).
- **Lynch syndrome** is caused by inherited mutations in genes responsible for **DNA mismatch repair**, leading to an accumulation of errors and increased cancer risk.
*Homologous recombination*
- This repair mechanism is crucial for fixing **double-strand DNA breaks** using a homologous DNA template, important for genetic stability and primarily associated with genes like BRCA1/2.
- While defects in homologous recombination can lead to cancer (e.g., **breast and ovarian cancers**), it is not the primary mechanism implicated in Lynch syndrome or the patient's specific presentation of colorectal and endometrial cancer families.
*Non-homologous end joining*
- This is another major pathway for repairing **double-strand DNA breaks**, but it does so by directly ligating the broken ends, often with some loss of genetic information, and does not rely on a homologous template.
- Defects in non-homologous end joining are not typically linked to the specific spectrum of cancers seen in **Lynch syndrome**.
*Nucleotide excision repair*
- This pathway is responsible for removing bulky DNA lesions, such as those caused by **UV light (e.g., pyrimidine dimers)** or certain chemical mutagens, and its defects are associated with conditions like xeroderma pigmentosum.
- The clinical picture and family history are not characteristic of disorders related to impaired **nucleotide excision repair**.
*Base excision repair*
- This repair pathway primarily corrects small, non-bulky DNA lesions, such as **oxidized, alkylated, or deaminated bases**, that do not distort the DNA helix.
- While important for maintaining genomic integrity, defects in base excision repair are typically associated with different cancer susceptibilities and not the specific features of **Lynch syndrome**.
Base excision repair US Medical PG Question 9: An investigator studying DNA mutation mechanisms isolates single-stranded DNA from a recombinant bacteriophage and sequences it. The investigator then mixes it with a buffer solution and incubates the resulting mixture at 70°C for 16 hours. Subsequent DNA resequencing shows that 3.7 per 1,000 cytosine residues have mutated to uracil. Which of the following best describes the role of the enzyme that is responsible for the initial step in repairing these types of mutations in living cells?
- A. Connecting the phosphodiester backbone
- B. Cleavage of the phosphodiester bond 3' of damaged site
- C. Creation of abasic site (Correct Answer)
- D. Release of the damaged nucleotide
- E. Addition of free nucleotides to 3' end
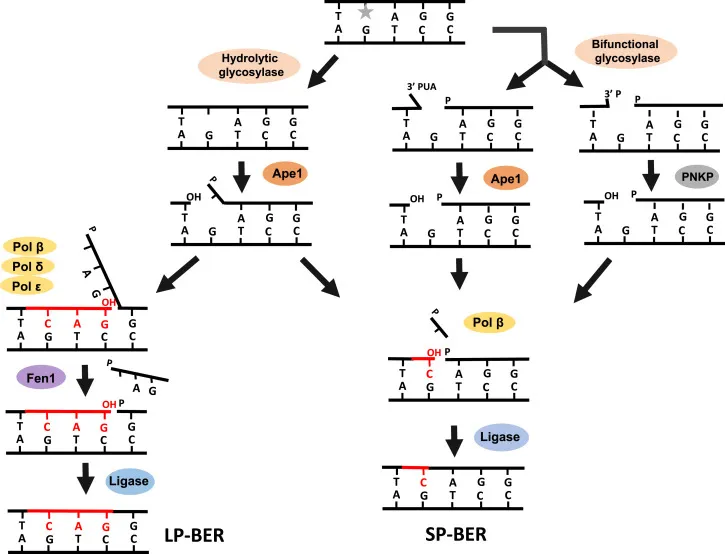
Base excision repair Explanation: ***Creation of abasic site***
- The mutation of **cytosine to uracil** is an example of **deamination**, which is repaired by the **base excision repair (BER)** pathway.
- The initial step in BER involves **DNA glycosylase**, which *removes* the damaged base (uracil) from the sugar-phosphate backbone by hydrolyzing the **N-glycosidic bond**, creating an **abasic site**.
*Connecting the phosphodiester backbone*
- This is the function of **DNA ligase**, which acts at the *final step* of DNA repair pathways to seal the nicks in the backbone.
- It does not initiate the repair process for deaminated bases.
*Cleavage of the phosphodiester bond 3' of damaged site*
- This is typically performed by an **AP endonuclease (APE1)** after the abasic site has been created.
- It is a *subsequent step* in BER, not the initial one for removing the damaged base itself.
*Release of the damaged nucleotide*
- While the damaged base is eventually *released*, the initial enzyme (DNA glycosylase) specifically removes the **base**, leaving the sugar and phosphate intact.
- The entire nucleotide (base, sugar, and phosphate) is typically removed later by an **AP lyase** or APE1, after the initial glycosylase action.
*Addition of free nucleotides to 3' end*
- This is the function of **DNA polymerase**, which fills in the gap after the damaged nucleotide and surrounding region have been excised.
- This occurs *after* the initial recognition and removal of the damaged base, not as the primary repair step.
Base excision repair US Medical PG Question 10: A previously healthy 42-year-old man comes to the emergency room with constipation and diffuse, worsening abdominal pain for 2 days. He has no history of major medical illness. His father died in a car accident at the age of 32 years, and his mother has type 2 diabetes mellitus. A diagnosis of bowel obstruction is suspected and he is taken to the operating room for exploratory laparotomy. A partial resection of the colon is performed. The gross appearance of the patient's colonic tissue is shown. Microscopic examination shows tubular, tubulovillous, and villous adenomas. Assuming the patient's partner is not a carrier of the condition, which of the following is the likelihood that this patient’s children will develop this condition?
- A. 50% (Correct Answer)
- B. 25%
- C. 100%
- D. 0%
- E. 75%
Base excision repair Explanation: ***50%***
- The image and clinical scenario are highly suggestive of **familial adenomatous polyposis (FAP)**, an autosomal dominant condition characterized by hundreds to thousands of colonic adenomas.
- Since FAP is an **autosomal dominant** disorder, an affected individual (heterozygous for the gene) has a **50% chance** of passing the mutated gene to each child, regardless of whether the partner is a carrier.
*25%*
- A 25% likelihood would be expected in **autosomal recessive inheritance** if both parents are carriers.
- FAP does not follow an autosomal recessive inheritance pattern; it is an **autosomal dominant** disease.
*100%*
- A 100% likelihood would occur only if the affected parent were **homozygous for the mutation** (extremely rare and typically lethal) or in certain non-Mendelian inheritance patterns.
- In typical FAP cases, the affected parent is heterozygous, resulting in a 50% chance of transmission to offspring, not 100%.
*0%*
- A 0% likelihood would imply that the condition is not hereditary or that the affected parent cannot transmit the disease, which is incorrect for FAP.
- FAP is a **hereditary condition** with a clear autosomal dominant inheritance pattern, so there is always a 50% risk of transmission per child.
*75%*
- A 75% likelihood does not correspond to any standard Mendelian inheritance pattern.
- This percentage does not fit the **autosomal dominant pattern** observed in FAP, which consistently shows 50% transmission risk per child.
More Base excision repair US Medical PG questions available in the OnCourse app. Practice MCQs, flashcards, and get detailed explanations.