DNA repair

On this page
🔬 The Cellular Guardian Network: DNA Repair Mastery
Every cell in your body sustains thousands of DNA lesions daily from metabolism, radiation, and replication errors, yet most go unnoticed because sophisticated repair systems constantly patrol your genome. You'll discover how cells detect damage through molecular sensors, triage lesions to specialized repair pathways, and execute precise corrections-from single-base edits to double-strand break reconstruction. Understanding these mechanisms reveals why some cancers resist chemotherapy, how inherited repair defects cause disease, and where targeted therapies can exploit repair vulnerabilities to selectively kill malignant cells.
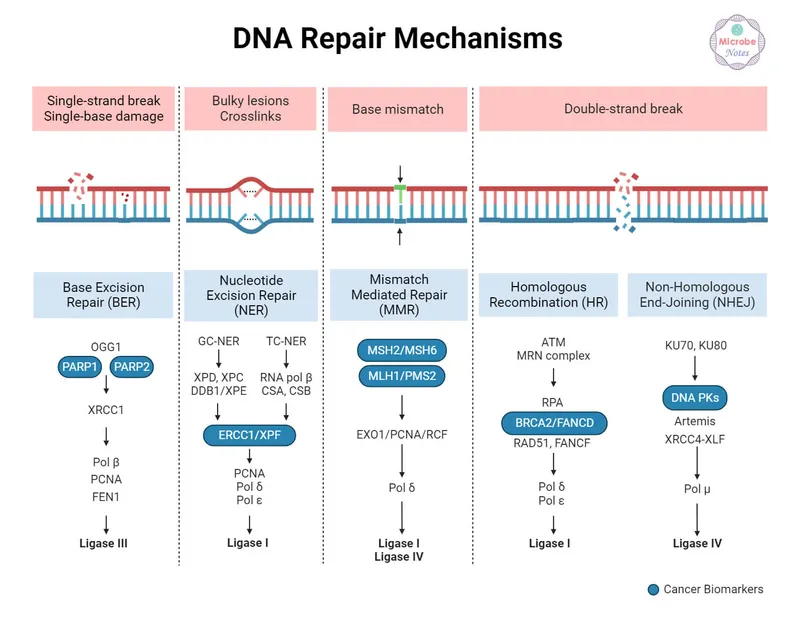
DNA repair represents the most sophisticated quality control system in biology, operating through five major pathways that collectively maintain genomic stability. Each pathway targets specific damage types with >99% efficiency, preventing the 10,000+ DNA lesions that occur daily in every human cell from becoming permanent mutations.
📌 Remember: BENDER - Base excision repair (oxidative damage), Excision repair nucleotide (bulky lesions), Non-homologous end joining (double breaks), Direct repair (alkylation), Error correction mismatch (replication errors), Recombination homologous (sister chromatid repair)
The repair machinery operates with extraordinary precision, distinguishing between normal DNA variations and pathological damage through molecular recognition systems that detect structural distortions as small as 0.1 nanometers. This surveillance network processes damage signals through ATM/ATR kinase cascades within minutes of detection, coordinating repair responses that can halt cell division, recruit specific repair proteins, or trigger apoptosis when damage exceeds repair capacity.
-
Spontaneous Damage Sources (daily cellular assault)
- Hydrolytic reactions: 10,000 base modifications/cell/day
- Oxidative stress: 2,000 8-oxoguanine lesions/cell/day
- Replication errors: 1 error per 10^10 nucleotides (post-proofreading)
- Mismatch frequency: 1:1000 before MMR correction
- Slippage events: 1:100,000 in repetitive sequences
-
Environmental Damage Amplifiers
- UV radiation: 100,000+ pyrimidine dimers/cell (1 hour sun exposure)
- Ionizing radiation: 40 double-strand breaks/Gy/cell
- Chemical mutagens: 1,000-fold increase in alkylation damage
- Cigarette smoke: 4,000+ DNA adducts/pack
- Alcohol metabolism: 500+ acetaldehyde crosslinks/drink
| Repair Pathway | Target Damage | Detection Time | Repair Efficiency | Clinical Deficiency | Cancer Risk Increase |
|---|---|---|---|---|---|
| Base Excision Repair | Oxidative, alkylation | <5 minutes | 99.9% | MUTYH syndrome | 100-fold colorectal |
| Nucleotide Excision | UV dimers, bulky adducts | 15-30 minutes | 95% | Xeroderma pigmentosum | 1000-fold skin cancer |
| Mismatch Repair | Replication errors | 2-4 hours | 99% | Lynch syndrome | 80% lifetime colorectal |
| Homologous Recombination | Double-strand breaks | 1-6 hours | 90% | BRCA mutations | 70% breast/ovarian |
| Non-homologous End Joining | Double-strand breaks | 30 minutes | 85% | SCID syndromes | Lymphoma/leukemia |
💡 Master This: DNA repair deficiency creates mutator phenotypes where mutation rates increase 100-10,000 fold, driving cancer development through genomic instability rather than single oncogene activation - explaining why repair-deficient cancers often respond better to DNA-damaging chemotherapy despite seeming paradoxical.
Understanding repair pathway coordination reveals how cells prioritize error-free mechanisms during S/G2 phases when sister chromatids are available, while relying on error-prone NHEJ during G1 phase when homologous templates are absent, creating cell cycle-dependent vulnerability windows for targeted cancer therapy.
🔬 The Cellular Guardian Network: DNA Repair Mastery
⚡ Damage Detection Command Center: Molecular Surveillance Systems
📌 Remember: SHARP sensors - Single-strand breaks (PARP), Homologous recombination blocks (ATR), Asymmetric DNA ends (ATM), Replication fork stalling (ATR), Pyrimidine dimers (specialized sensors)
-
ATM Kinase Cascade (double-strand break response)
- Activation threshold: 1-2 DSBs per cell
- Autophosphorylation sites: Ser1981, Ser367, Ser1893
- Downstream targets: >1000 proteins within 30 minutes
- p53 phosphorylation: Ser15 (transcriptional activation)
- Chk2 activation: Thr68 (cell cycle checkpoint)
- BRCA1 recruitment: Ser1524 (repair focus formation)
-
ATR Signaling Network (replication stress response)
- RPA-coated ssDNA threshold: >30 nucleotides
- ATRIP recruitment: <2 minutes post-damage
- Chk1 phosphorylation: Ser345/Ser317 (S-phase arrest)
- Fork protection: 70% reduction in collapse rate
- Origin firing suppression: 80% decrease in late origins
- Dormant origin activation: 5-fold increase in backup firing
| Sensor Protein | Damage Recognition | Activation Time | Phosphorylation Targets | Deficiency Syndrome | Cellular Phenotype |
|---|---|---|---|---|---|
| ATM | DSB ends, chromatin breaks | 1-5 minutes | >1000 substrates | Ataxia-telangiectasia | Radiosensitivity, cancer |
| ATR | RPA-ssDNA, stalled forks | 2-10 minutes | >300 substrates | Seckel syndrome | Replication defects |
| PARP1 | SSBs, base damage | <30 seconds | >100 substrates | Rare variants | Mild repair defects |
| DNA-PKcs | DSB ends (NHEJ) | 1-3 minutes | >200 substrates | SCID | Immunodeficiency |
| p53 | Multiple inputs | 15-60 minutes | >500 targets | Li-Fraumeni | Cancer predisposition |
💡 Master This: Damage sensor activation creates positive feedback loops where initial repair attempts generate additional DNA breaks (e.g., nucleotide excision repair creates transient SSBs), requiring temporal coordination of sensor responses to prevent hyperactivation and inappropriate apoptosis induction.
The sensor network demonstrates remarkable damage threshold sensitivity, where single DSBs can trigger ATM activation affecting thousands of downstream targets, while chronic low-level damage from oxidative stress requires cumulative sensor input over hours to days before triggering permanent cell cycle arrest or senescence pathways.
⚡ Damage Detection Command Center: Molecular Surveillance Systems
🛠️ Repair Pathway Selection: The Cellular Triage System
Repair pathway selection operates through hierarchical decision trees where damage structure provides the primary sorting signal, cell cycle phase determines template availability, and chromatin modifications influence repair factor accessibility. This triage system prioritizes high-fidelity repair when possible while maintaining backup pathways for emergency situations.
-
Damage-Specific Pathway Routing
- Single base modifications: BER pathway (>95% efficiency)
- Bulky DNA adducts: NER pathway (15-30 nucleotide patches)
- Replication errors: MMR system (1000-fold error reduction)
- Mismatch recognition: MSH2/MSH6 (base-base mismatches)
- Insertion/deletion loops: MSH2/MSH3 (1-4 nucleotide slips)
- Strand discrimination: PCNA/RFC (newly synthesized strand)
-
Cell Cycle-Dependent Repair Choice
- G1 phase: NHEJ preferred (80% of DSB repair)
- No sister chromatid template available
- Ku70/Ku80 binding within seconds
- DNA-PKcs recruitment and autophosphorylation
- S/G2 phases: HR pathway dominant (70% of DSB repair)
- Sister chromatid template present
- CtIP-mediated end resection
- RAD51 nucleofilament formation
- G1 phase: NHEJ preferred (80% of DSB repair)
📌 Remember: CHOICE factors - Cell cycle phase (G1=NHEJ, S/G2=HR), Homology availability (sister chromatids), Oxidative damage (BER), Insertion/deletion errors (MMR), Chromatin state (open=accessible), End processing requirements (blunt vs. processed)
| Repair Decision Factor | Pathway Influence | Timing Impact | Fidelity Outcome | Clinical Relevance | Therapeutic Target |
|---|---|---|---|---|---|
| Cell Cycle Phase | G1→NHEJ, S/G2→HR | Immediate | HR>NHEJ accuracy | Cell cycle checkpoints | CDK inhibitors |
| Chromatin State | Open→accessible | 5-15 minutes | Context-dependent | Epigenetic therapy | HDAC inhibitors |
| Damage Complexity | Simple→fast, complex→slow | Minutes to hours | Inversely related | Radiation sensitivity | Radiosensitizers |
| Repair Factor Availability | Pathway competition | Variable | Resource-limited | Synthetic lethality | PARP inhibitors |
| Metabolic State | Energy-dependent | ATP/NAD+ levels | Quality control | Cancer metabolism | Metabolic inhibitors |
The repair selection system demonstrates pathway redundancy where multiple mechanisms can address similar damage types with different fidelity levels. For example, double-strand breaks can be repaired by HR (>99% accuracy), NHEJ (85-95% accuracy), or alternative end-joining (<50% accuracy), with pathway choice determining mutation frequency and chromosomal stability.
💡 Master This: Repair pathway selection creates therapeutic vulnerabilities where pathway-specific inhibitors can force cells into suboptimal repair modes, increasing mutation burden and genomic instability that preferentially affects rapidly dividing cancer cells over quiescent normal tissues.
Understanding pathway hierarchy reveals why combination therapies targeting multiple repair pathways simultaneously achieve synergistic effects, as cells cannot compensate for simultaneous pathway blockade and undergo catastrophic genomic instability leading to apoptosis or mitotic catastrophe.
🛠️ Repair Pathway Selection: The Cellular Triage System
🔍 Repair Mechanism Precision: Molecular Surgery Techniques
-
Base Excision Repair Precision
- Damage recognition: 8-oxoguanine detection by OGG1 glycosylase
- Base removal: N-glycosidic bond hydrolysis (<1 second)
- AP site processing: APE1 endonuclease creates 3'-OH and 5'-dRP
- Short-patch repair: 1 nucleotide replacement (80% of events)
- Long-patch repair: 2-10 nucleotides (20% of events)
- DNA ligase III sealing: ATP-dependent phosphodiester formation
-
Nucleotide Excision Repair Architecture
- Damage verification: XPA protein confirms helix distortion
- Dual incision: XPF-ERCC1 (3' cut) and XPG (5' cut)
- Oligonucleotide removal: 24-32 nucleotide patch excision
- TFIIH helicase: ATP-dependent unwinding (3-5 bp/second)
- RPA coating: ssDNA protection during gap filling
- PCNA-dependent synthesis: high-fidelity Pol δ/ε
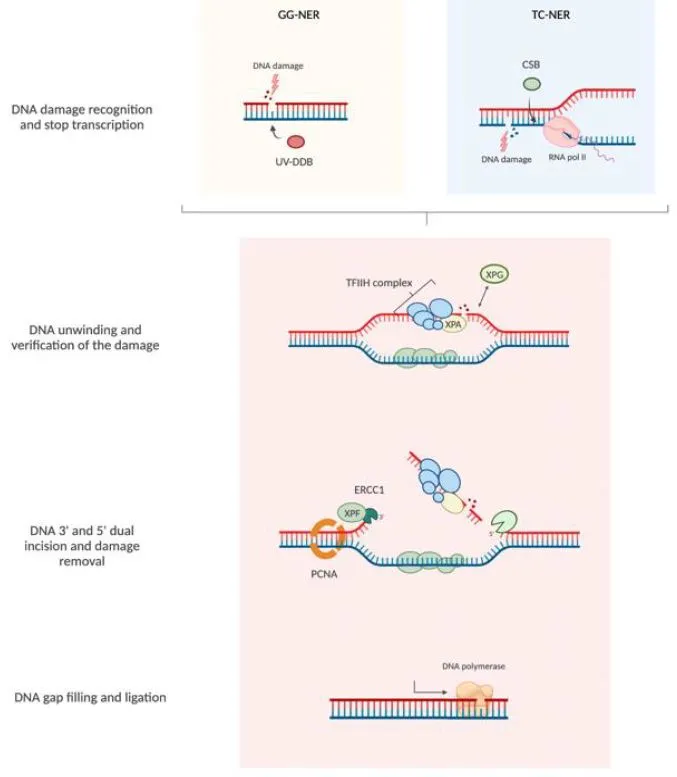
| Repair Enzyme | Substrate Specificity | Catalytic Rate | Error Frequency | Deficiency Disease | Functional Consequence |
|---|---|---|---|---|---|
| OGG1 | 8-oxoguanine | 0.1/second | <1:10^6 | Mild cancer risk | Oxidative damage accumulation |
| UNG | Uracil in DNA | 1/second | <1:10^7 | Immunodeficiency | Somatic hypermutation defects |
| XPG | 5' incision (NER) | 0.01/second | <1:10^5 | Xeroderma pigmentosum | UV sensitivity, cancer |
| MSH2 | Mismatch recognition | 0.1/second | 1:10^3 detection | Lynch syndrome | Microsatellite instability |
| PARP1 | SSB detection | 10/second | N/A (sensor) | Rare variants | Repair coordination defects |
⭐ Clinical Pearl: XP patients with XPG mutations show 1000-fold increased skin cancer risk due to defective 5' incision in nucleotide excision repair, requiring complete UV avoidance and prophylactic skin surveillance every 3-6 months to detect early malignancies.
The precision of repair mechanisms extends to chromatin restoration, where histone modifications are faithfully restored post-repair through chromatin remodeling complexes and histone chaperones that maintain epigenetic information across repair events, ensuring gene expression patterns remain unchanged despite DNA synthesis.
💡 Master This: Repair precision creates therapeutic windows where repair inhibitors can selectively target cancer cells with defective repair pathways while sparing normal cells with intact backup mechanisms, forming the basis for synthetic lethality approaches in precision oncology.
Understanding enzymatic precision reveals why repair pathway mutations often show tissue-specific phenotypes - UV-sensitive skin in XP, neurodegeneration in ataxia-telangiectasia, immunodeficiency in SCID - reflecting tissue-specific repair demands and damage exposure patterns that exceed residual repair capacity.
🔍 Repair Mechanism Precision: Molecular Surgery Techniques
⚖️ Therapeutic Targeting: Exploiting Repair Vulnerabilities
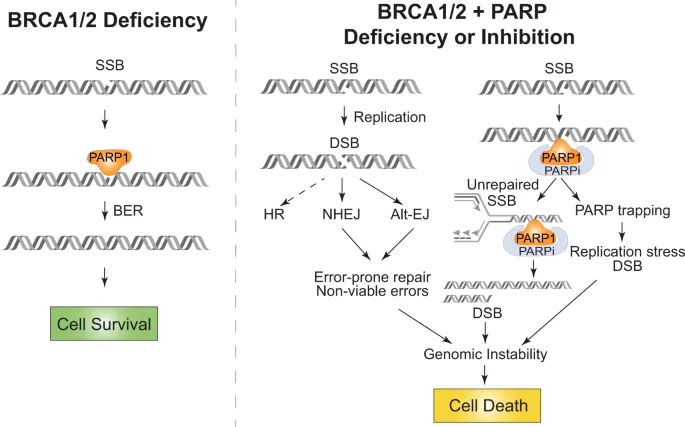
Therapeutic targeting leverages repair pathway dependencies where cancer cells with defective repair mechanisms become hyperdependent on remaining functional pathways. PARP inhibitors exemplify this approach, achieving 60-80% response rates in BRCA-mutated ovarian cancers by overwhelming residual repair capacity and forcing catastrophic genomic instability.
-
PARP Inhibitor Mechanisms
- PARP1/2 trapping: DNA-protein crosslink formation
- Base excision repair blockade: SSB accumulation
- Replication fork collision: DSB conversion during S-phase
- Olaparib: IC50 = 5nM (PARP1), FDA-approved 2014
- Rucaparib: IC50 = 1.4nM (PARP1), platinum-sensitive recurrence
- Niraparib: IC50 = 3.8nM (PARP1), maintenance therapy
-
Combination Therapy Strategies
- PARP + Checkpoint inhibitors: 70% response in BRCA-mutated cancers
- PARP + ATR inhibitors: Synthetic lethality in HR-deficient tumors
- PARP + DNA damaging agents: Radiosensitization (2-5 fold)
- Cisplatin + Olaparib: Progression-free survival 19.1 months
- Radiation + PARP inhibition: Enhancement ratio 1.5-2.0
- Temozolomide + PARP: MGMT-deficient glioblastoma targeting
| Therapeutic Target | Mechanism | Cancer Type | Response Rate | Resistance Mechanism | Combination Strategy |
|---|---|---|---|---|---|
| PARP1/2 | SSB repair blockade | BRCA-mutated | 60-80% | HR restoration | ATR inhibitors |
| ATR kinase | Replication checkpoint | Replication stress | 30-50% | p53 activation | PARP inhibitors |
| CHK1 kinase | S-phase checkpoint | p53-deficient | 20-40% | Checkpoint adaptation | Gemcitabine |
| DNA-PKcs | NHEJ inhibition | HR-deficient | 40-60% | Alternative end-joining | Radiation |
| WEE1 kinase | G2/M checkpoint | p53-mutated | 25-45% | CDK1 mutations | DNA damaging agents |
⭐ Clinical Pearl: Platinum-resistant ovarian cancers often develop secondary BRCA mutations that restore reading frame and HR function, conferring PARP inhibitor resistance. Circulating tumor DNA monitoring can detect these reversion mutations months before clinical progression, guiding therapy switching.
The therapeutic landscape continues expanding with next-generation inhibitors targeting repair pathway crosstalk: ATR inhibitors show synergy with PARP inhibitors in HR-proficient cancers, CHK1 inhibitors enhance replication stress in p53-deficient tumors, and DNA-PKcs inhibitors sensitize radioresistant cancers to ionizing radiation.
💡 Master This: Successful repair-targeted therapy requires biomarker-driven patient selection using homologous recombination deficiency scores, genomic instability signatures, and functional repair assays to identify patients most likely to benefit from specific repair inhibitor combinations.
Understanding resistance mechanisms reveals evolutionary pressure where surviving cancer cells develop compensatory mutations, pathway rewiring, or drug efflux mechanisms, necessitating combination approaches that target multiple repair pathways simultaneously to prevent adaptive resistance and achieve durable responses.
⚖️ Therapeutic Targeting: Exploiting Repair Vulnerabilities
🔗 Systems Integration: The Repair Network Ecosystem
-
Pathway Crosstalk Networks
- BER-NER coordination: PARP1 signals recruit NER factors for complex lesions
- HR-NHEJ competition: CtIP versus 53BP1 determines pathway choice
- MMR-HR integration: MSH2/MSH6 can trigger HR at replication forks
- Shared factors: PCNA coordinates replication and repair
- RPA protein: Universal ssDNA coating in multiple pathways
- ATM/ATR signaling: Checkpoint coordination across all pathways
-
Metabolic Integration Points
- NAD+ consumption: PARP activation depletes cellular NAD+ pools
- ATP requirements: Helicase activity demands high energy input
- dNTP availability: Repair synthesis competes with replication
- Ribonucleotide reductase: Rate-limiting for dNTP production
- Thymidine kinase: Salvage pathway during repair stress
- SAMHD1 regulation: dNTP pool homeostasis maintenance
| Integration Level | Key Components | Coordination Mechanism | Failure Consequence | Clinical Manifestation | Therapeutic Opportunity |
|---|---|---|---|---|---|
| Pathway Crosstalk | Shared proteins, signals | Competitive binding | Repair pathway imbalance | Cancer predisposition | Pathway-specific targeting |
| Cell Cycle Integration | Checkpoints, CDKs | Phosphorylation cascades | Genomic instability | Developmental disorders | Cell cycle inhibitors |
| Metabolic Coupling | NAD+, ATP, dNTPs | Resource competition | Energy-dependent failure | Metabolic syndromes | Metabolic modulators |
| Stress Response | p53, NF-κB pathways | Transcriptional programs | Inappropriate cell death | Tissue degeneration | Stress pathway drugs |
| Epigenetic Control | Chromatin modifiers | Histone modifications | Gene expression changes | Developmental defects | Epigenetic therapies |
⭐ Clinical Pearl: Fanconi anemia patients demonstrate network-wide repair failure affecting multiple pathways simultaneously, requiring comprehensive management including bone marrow transplantation (90% cure rate), cancer surveillance (lifetime risk >50%), and fertility preservation due to gonadal dysfunction.
The network architecture reveals emergent properties where system-level behaviors exceed individual pathway capabilities. Repair pathway redundancy provides robustness against single pathway failures, while pathway competition can create vulnerabilities when multiple systems are simultaneously compromised.
💡 Master This: Network-level understanding enables precision medicine approaches where multi-pathway biomarkers predict therapeutic responses better than single gene mutations, guiding combination therapy selection and resistance monitoring through systems-level analysis.
Integration complexity explains why repair disorders often present with multi-system phenotypes - neurodegeneration, immunodeficiency, cancer predisposition, and developmental abnormalities - reflecting tissue-specific repair demands and network vulnerability patterns that vary across cell types and developmental stages.
🔗 Systems Integration: The Repair Network Ecosystem
🎯 Clinical Mastery Arsenal: Rapid-Fire Repair Diagnostics
Master clinicians recognize repair deficiency patterns through constellation findings that combine cancer types, age of onset, family clustering, and treatment responses into diagnostic signatures that guide genetic testing, surveillance protocols, and therapeutic selection with evidence-based precision.
📌 Remember: REPAIR clinical signs - Recurrent cancers (multiple primaries), Early onset (<50 years), Pedigree clustering (family history), Atypical presentations (rare sites), Immune dysfunction (infections), Radiation sensitivity (severe reactions)
-
Essential Clinical Thresholds
- Lynch syndrome: >70% lifetime colorectal cancer risk
- BRCA mutations: 60-80% breast, 40-60% ovarian cancer risk
- Li-Fraumeni: >90% lifetime cancer risk (any type)
- Radiation sensitivity: 50-70% dose reduction required
- Chemotherapy toxicity: Grade 3-4 reactions in >80%
- Surgical considerations: Prophylactic procedures reduce risk >90%
-
Rapid Diagnostic Framework
- Age <50 + colorectal cancer → Lynch syndrome testing
- Triple-negative breast + family history → BRCA analysis
- Multiple primaries + radiation sensitivity → ATM evaluation
- Microsatellite instability: >95% sensitivity for MMR defects
- Tumor mutational burden: >10 mutations/Mb suggests repair deficiency
- Homologous recombination deficiency score: >42 predicts PARP sensitivity
| Clinical Scenario | Genetic Test Priority | Surveillance Protocol | Treatment Modification | Risk Reduction Strategy | Family Screening |
|---|---|---|---|---|---|
| Early-onset colorectal | Lynch panel (MMR genes) | Colonoscopy q1-2 years | MSI-high = immunotherapy | Prophylactic surgery | First-degree relatives |
| Triple-negative breast <40 | BRCA1/2 sequencing | MRI + mammography q6mo | PARP inhibitor eligible | Bilateral mastectomy | Cascade testing |
| Radiation hypersensitivity | ATM sequencing | Enhanced imaging surveillance | 50% dose reduction | Avoid radiation therapy | Clinical assessment |
| Multiple primary cancers | Comprehensive panel | Multi-organ screening | Targeted therapy selection | Risk-reducing surgery | Genetic counseling |
| Childhood cancer + family Hx | Li-Fraumeni (p53) | Whole-body MRI annually | Avoid radiation | Intensive surveillance | Predictive testing |
💡 Master This: Tumor-first genetic testing approaches identify somatic repair defects in 15-20% of cancers without germline mutations, expanding targeted therapy eligibility and revealing acquired vulnerabilities that guide precision treatment selection beyond hereditary cancer syndromes.
The clinical arsenal includes functional assays that measure actual repair capacity rather than genetic status alone: RAD51 focus formation assays assess HR function, comet assays measure overall repair efficiency, and chromosomal instability analysis reveals pathway-specific defects that predict therapeutic responses with greater accuracy than mutation testing alone.
Understanding repair deficiency presentations enables proactive management where early detection, risk-reducing interventions, and targeted therapies transform high-risk genetic profiles into manageable clinical conditions with significantly improved outcomes through precision medicine approaches tailored to individual repair pathway status.
🎯 Clinical Mastery Arsenal: Rapid-Fire Repair Diagnostics
Practice Questions: DNA repair
Test your understanding with these related questions
A 5-month-old male infant from a consanguineous marriage presents with severe sunburns and freckling in sun exposed areas. The mother explains that the infant experiences these sunburns every time the infant goes outside despite applying copious amounts of sunscreen. Which of the following DNA repair mechanisms is defective in this child?












